Marked neurotropism and potential adaptation of H5N1 clade 2.3.4.4.b virus in naturally infected domestic cats
- PMID: 39648950
- PMCID: PMC11654043
- DOI: 10.1080/22221751.2024.2440498
Marked neurotropism and potential adaptation of H5N1 clade 2.3.4.4.b virus in naturally infected domestic cats
Abstract
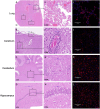
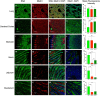
In April 2024, ten cats died in a rural South Dakota (SD) residence, showing respiratory and neurological symptoms. Necropsy and laboratory testing of two cats confirmed H5N1 clade 2.3.4.4b infection. The viral genome sequences are closely related to recent SD cattle H5N1 sequences. Cat H5N1 genomes had unique mutations, including T143A in haemagglutinin, known to affect infectivity and immune evasion, and two novel mutations in PA protein (F314L, L342Q) that may affect polymerase activity and virulence, suggesting potential virus adaptation. Dead cats showed systemic infection with lesions and viral antigens in multiple organs. Higher viral RNA and antigen in the brain indicated pronounced neurotropism. Lectin-histochemistry revealed widespread co-expression of sialic acid α-2,6 and α-2,3 receptors, suggesting cats could serve as mixing vessels for reassortment of avian and mammalian influenza viruses. No differences in clade 2.2 or 2.3.4.4b H5 pseudoviruses binding to cat lung/brain tissues indicated the neurotropism is unlikely mediated by receptor binding affinity.
Keywords: A(H5N1); Influenza A viruses; avian influenza; cat; clade 2.3.4.4b; influenza A virus evolution; neurotropism.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Blushke C. Influenza A (H5N1) virus detected in Wild Foxes (Vulpes vulpes) in Ontario. Healthy Wildlife, The blog of the Canadian Wildlife Health Cooperative, 2022.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
