This is a preprint.
Multi-ancestry genome-wide association study reveals novel genetic signals for lung function decline
- PMID: 39649580
- PMCID: PMC11623738
- DOI: 10.1101/2024.11.25.24317794
Multi-ancestry genome-wide association study reveals novel genetic signals for lung function decline
Abstract
Rationale: Accelerated decline in lung function contributes to the development of chronic respiratory disease. Despite evidence for a genetic component, few genetic associations with lung function decline have been identified.
Objectives: To evaluate genome-wide associations and putative downstream functionality of genetic variants with lung function decline in diverse general population cohorts.
Methods: We conducted genome-wide association study (GWAS) analyses of decline in the forced expiratory volume in the first second (FEV1), forced vital capacity (FVC), and their ratio (FEV1/FVC) in participants across six cohorts in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium and the UK Biobank. Genotypes were imputed to TOPMed (CHARGE cohorts) or Haplotype Reference Consortium (HRC) (UK Biobank) reference panels, and GWAS analyses used generalized estimating equation models with robust standard error. Models were stratified by cohort, ancestry, and sex, and adjusted for important lung function confounders and genotype principal components. Results were combined in cross-ancestry and ancestry-specific meta-analyses. Selected top variants were tested for replication in two independent COPD-enriched cohorts.
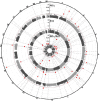
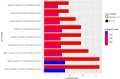
Measurements and main results: Our discovery analyses included 52,056 self-reported White (N=44,988), Black (N=5,788), Hispanic (N=550), and Chinese American (N=730) participants with a mean of 2.3 spirometry measurements and 8.6 years of follow-up. Functional mapping of GWAS meta-analysis results identified 361 distinct genome-wide significant (p<5E-08) variants in one or more of the FEV1, FVC, and FEV1/FVC decline phenotypes, which overlapped with previously reported genetic signals for several related pulmonary traits. Of these, 8 variants, or 20.5% of the variant set available for replication testing, were nominally associated (p<0.05) with at least one decline phenotype in COPD-enriched cohorts (White [N=4,778] and Black [N=1,118]). Using the GWAS results, gene-level analysis implicated 38 genes, including eight (XIRP2, GRIN2D, SATB1, MARCHF4, SIPA1L2, ANO5, H2BC10, and FAF2) with consistent associations across ancestries or decline phenotypes. Annotation class analysis revealed significant enrichment of several regulatory processes for corticosteroid biosynthesis and metabolism. Drug repurposing analysis identified 43 approved compounds targeting eight of the implicated 38 genes.
Conclusions: Our multi-ancestry GWAS meta-analyses identified numerous genetic loci associated with lung function decline. These findings contribute knowledge to the genetic architecture of lung function decline, provide evidence for a role of endogenous corticosteroids in the etiology of lung function decline, and identify drug targets that merit further study for potential repurposing to slow lung function decline and treat lung disease.
Keywords: Chronic Obstructive; Genomics; Lung Volume Measurements; Pulmonary Disease; Spirometry.
Conflict of interest statement
COMPETING INTERESTS BMP serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. No other others have competing interests to declare.
Figures



References
-
- Shrine N., Guyatt A.L., Erzurumluoglu A.M., Jackson V.E., Hobbs B.D., Melbourne C.A., Batini C., Fawcett K.A., Song K., Sakornsakolpat P., et al. (2019). New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat. Genet. 51, 481–493. 10.1038/s41588-018-0321-7. - DOI - PMC - PubMed
-
- Shrine N., Izquierdo A.G., Chen J., Packer R., Hall R.J., Guyatt A.L., Batini C., Thompson R.J., Pavuluri C., Malik V., et al. (2023). Multi-ancestry genome-wide association analyses improve resolution of genes and pathways influencing lung function and chronic obstructive pulmonary disease risk. Nat. Genet. 55, 410–422. 10.1038/s41588-023-01314-0. - DOI - PMC - PubMed
-
- Wain L.V., Shrine N., Artigas M.S., Erzurumluoglu A.M., Noyvert B., Bossini-Castillo L., Obeidat M., Henry A.P., Portelli M.A., Hall R.J., et al. (2017). Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat. Genet. 49, 416–425. 10.1038/ng.3787. - DOI - PMC - PubMed
Publication types
Grants and funding
- U01 HL089897/HL/NHLBI NIH HHS/United States
- N01 HC048050/HL/NHLBI NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- N01 HC095160/HL/NHLBI NIH HHS/United States
- N01 HC095095/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- R01 HL103612/HL/NHLBI NIH HHS/United States
- 75N92020D00002/HL/NHLBI NIH HHS/United States
- N01 HC095161/HL/NHLBI NIH HHS/United States
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- N01 HC048048/HL/NHLBI NIH HHS/United States
- R21 HL153700/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N02 HL064278/HL/NHLBI NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- 75N92020D00001/HL/NHLBI NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- N01 HC048049/HL/NHLBI NIH HHS/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- R01 HL089856/HL/NHLBI NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- R01 NR012459/NR/NINR NIH HHS/United States
- 75N92020D00003/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- HHSN268201700002C/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- R21 HL129924/HL/NHLBI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- 75N92020D00004/HL/NHLBI NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- 75N92020D00007/HL/NHLBI NIH HHS/United States
- N01 HC048047/HL/NHLBI NIH HHS/United States
- R01 AG028050/AG/NIA NIH HHS/United States
- HHSN268201500014C/HL/NHLBI NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- 75N92021D00006/HL/NHLBI NIH HHS/United States
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- 75N92019D00031/HL/NHLBI NIH HHS/United States
- HHSN268201700004C/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- N01 HC095166/HL/NHLBI NIH HHS/United States
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- 75N92020D00006/HL/NHLBI NIH HHS/United States
- K23 HL130627/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- R01 HL087641/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
