This is a preprint.
Blood methylation biomarkers are associated with diabetic kidney disease progression in type 1 diabetes
- PMID: 39649605
- PMCID: PMC11623717
- DOI: 10.1101/2024.11.28.24318055
Blood methylation biomarkers are associated with diabetic kidney disease progression in type 1 diabetes
Abstract
Background: DNA methylation differences are associated with kidney function and diabetic kidney disease (DKD), but prospective studies are scarce. Therefore, we aimed to study DNA methylation in a prospective setting in the Finnish Diabetic Nephropathy Study type 1 diabetes (T1D) cohort.
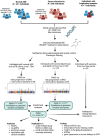
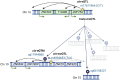
Methods: We analysed baseline blood sample-derived DNA methylation (Illumina's EPIC array) of 403 individuals with normal albumin excretion rate (early progression group) and 373 individuals with severe albuminuria (late progression group) and followed-up their DKD progression defined as decrease in eGFR to <60 mL/min/1.73m2 (early DKD progression group; median follow-up 13.1 years) or end-stage kidney disease (ESKD) (late DKD progression group; median follow-up 8.4 years). We conducted two epigenome-wide association studies (EWASs) on DKD progression and sought methylation quantitative trait loci (meQTLs) for the lead CpGs to estimate genetic contribution.
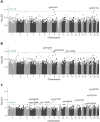
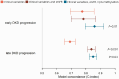
Results: Altogether, 14 methylation sites were associated with DKD progression (P<9.4×10-8). Methylation at cg01730944 near CDKN1C and at other CpGs associated with early DKD progression were not correlated with baseline eGFR, whereas late progression CpGs were strongly associated. Importantly, 13 of 14 CpGs could be linked to a gene showing differential expression in DKD or chronic kidney disease. Higher methylation at the lead CpG cg17944885, a frequent finding in eGFR EWASs, was associated with ESKD risk (HR [95% CI] = 2.15 [1.79, 2.58]). Additionally, we replicated meQTLs for cg17944885 and identified ten novel meQTL variants for other CpGs. Furthermore, survival models including the significant CpG sites showed increased predictive performance on top of clinical risk factors.
Conclusions: Our EWAS on early DKD progression identified a podocyte-specific CDKN1C locus. EWAS on late progression proposed novel CpGs for ESKD risk and confirmed previously known sites for kidney function. Since DNA methylation signals could improve disease course prediction, a combination of blood-derived methylation sites could serve as a potential prognostic biomarker.
Figures

References
-
- Jansson Sigfrids F, Groop PH, Harjutsalo V. Incidence rate patterns, cumulative incidence, and time trends for moderate and severe albuminuria in individuals diagnosed with type 1 diabetes aged 0-14 years: a population-based retrospective cohort study. Lancet Diabetes Endocrinol. 2022;10(7):489–498 doi:10.1016/S2213-8587(22)00099-7 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
