Substrate prediction for RiPP biosynthetic enzymes via masked language modeling and transfer learning
- PMID: 39649639
- PMCID: PMC11622008
- DOI: 10.1039/d4dd00170b
Substrate prediction for RiPP biosynthetic enzymes via masked language modeling and transfer learning
Abstract
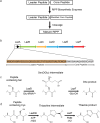
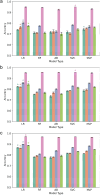
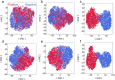
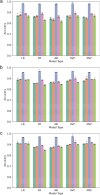
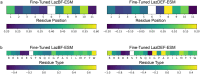
Ribosomally synthesized and post-translationally modified peptide (RiPP) biosynthetic enzymes often exhibit promiscuous substrate preferences that cannot be reduced to simple rules. Large language models are promising tools for predicting the specificity of RiPP biosynthetic enzymes. However, state-of-the-art protein language models are trained on relatively few peptide sequences. A previous study comprehensively profiled the peptide substrate preferences of LazBF (a two-component serine dehydratase) and LazDEF (a three-component azole synthetase) from the lactazole biosynthetic pathway. We demonstrated that masked language modeling of LazBF substrate preferences produced language model embeddings that improved downstream prediction of both LazBF and LazDEF substrates. Similarly, masked language modeling of LazDEF substrate preferences produced embeddings that improved prediction of both LazBF and LazDEF substrates. Our results suggest that the models learned functional forms that are transferable between distinct enzymatic transformations that act within the same biosynthetic pathway. We found that a single high-quality data set of substrates and non-substrates for a RiPP biosynthetic enzyme improved substrate prediction for distinct enzymes in data-scarce scenarios. We then fine-tuned models on each data set and showed that the fine-tuned models provided interpretable insight that we anticipate will facilitate the design of substrate libraries that are compatible with desired RiPP biosynthetic pathways.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Update of
-
Substrate Prediction for RiPP Biosynthetic Enzymes via Masked Language Modeling and Transfer Learning.ArXiv [Preprint]. 2024 Feb 23:arXiv:2402.15181v1. ArXiv. 2024. Update in: Digit Discov. 2024 Dec 2;4(2):343-354. doi: 10.1039/d4dd00170b. PMID: 38463513 Free PMC article. Updated. Preprint.
References
-
- Montalbán-López M. Scott T. A. Ramesh S. Rahman I. R. van Heel A. J. Viel J. H. Bandarian V. Dittmann E. Genilloud O. Goto Y. Burgos M. J. G. Hill C. Kim S. Koehnke J. Latham J. A. Link A. J. Martínez B. Nair S. K. Nicolet Y. Rebuffat S. Sahl H.-G. Sareen D. Schmidt E. W. Schmitt L. Severinov K. Süssmuth R. D. Truman A. W. Wang H. Weng J.-K. van Wezel G. P. Zhang Q. Zhong J. Piel J. Mitchell D. A. Kuipers O. P. van der Donk W. A. Nat. Prod. Rep. 2021;38:130–239. doi: 10.1039/D0NP00027B. - DOI - PMC - PubMed
-
- Arnison P. G. Bibb M. J. Bierbaum G. Bowers A. A. Bugni T. S. Bulaj G. Camarero J. A. Campopiano D. J. Challis G. L. Clardy J. Cotter P. D. Craik D. J. Dawson M. Dittmann E. Donadio S. Dorrestein P. C. Entian K.-D. Fischbach M. A. Garavelli J. S. Göransson U. Gruber C. W. Haft D. H. Hemscheidt T. K. Hertweck C. Hill C. Horswill A. R. Jaspars M. Kelly W. L. Klinman J. P. Kuipers O. P. Link A. J. Liu W. Marahiel M. A. Mitchell D. A. Moll G. N. Moore B. S. Müller R. Nair S. K. Nes I. F. Norris G. E. Olivera B. M. Onaka H. Patchett M. L. Piel J. Reaney M. J. T. Rebuffat S. Ross R. P. Sahl H.-G. Schmidt E. W. Selsted M. E. Severinov K. Shen B. Sivonen K. Smith L. Stein T. Süssmuth R. D. Tagg J. R. Tang G.-L. Truman A. W. Vederas J. C. Walsh C. T. Walton J. D. Wenzel S. C. Willey J. M. van der Donk W. A. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/C2NP20085F. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
