This is a preprint.
Design of high specificity binders for peptide-MHC-I complexes
- PMID: 39651227
- PMCID: PMC11623666
- DOI: 10.1101/2024.11.28.625793
Design of high specificity binders for peptide-MHC-I complexes
Update in
-
Design of high-specificity binders for peptide-MHC-I complexes.Science. 2025 Jul 24;389(6758):386-391. doi: 10.1126/science.adv0185. Epub 2025 Jul 24. Science. 2025. PMID: 40705892
Abstract
Class I MHC molecules present peptides derived from intracellular antigens on the cell surface for immune surveillance, and specific targeting of these peptide-MHC (pMHC) complexes could have considerable utility for treating diseases. Such targeting is challenging as it requires readout of the few outward facing peptide antigen residues and the avoidance of extensive contacts with the MHC carrier which is present on almost all cells. Here we describe the use of deep learning-based protein design tools to denovo design small proteins that arc above the peptide binding groove of pMHC complexes and make extensive contacts with the peptide. We identify specific binders for ten target pMHCs which when displayed on yeast bind the on-target pMHC tetramer but not closely related peptides. For five targets, incorporation of designs into chimeric antigen receptors leads to T-cell activation by the cognate pMHC complexes well above the background from complexes with peptides derived from proteome. Our approach can generate high specificity binders starting from either experimental or predicted structures of the target pMHC complexes, and should be widely useful for both protein and cell based pMHC targeting.
Figures
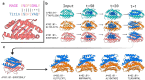


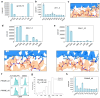
References
-
- Yarmarkovich M., Marshall Q. F., Warrington J. M., Premaratne R., Farrel A., Groff D., Li W., di Marco M., Runbeck E., Truong H., Toor J. S., Tripathi S., Nguyen S., Shen H., Noel T., Church N. L., Weiner A., Kendsersky N., Martinez D., Weisberg R., Christie M., Eisenlohr L., Bosse K. R., Dimitrov D. S., Stevanovic S., Sgourakis N. G., Kiefel B. R., Maris J. M., Targeting of intracellular oncoproteins with peptide-centric CARs. Nature 623, 820–827 (2023). - PMC - PubMed
-
- Hsiue E. H.-C., Wright K. M., Douglass J., Hwang M. S., Mog B. J., Pearlman A. H., Paul S., DiNapoli S. R., Konig M. F., Wang Q., Schaefer A., Miller M. S., Skora A. D., Azurmendi P. A., Murphy M. B., Liu Q., Watson E., Li Y., Pardoll D. M., Bettegowda C., Papadopoulos N., Kinzler K. W., Vogelstein B., Gabelli S. B., Zhou S., Targeting a neoantigen derived from a common TP53 mutation. Science 371 (2021). - PMC - PubMed
-
- Nathan P., Hassel J. C., Rutkowski P., Baurain J.-F., Butler M. O., Schlaak M., Sullivan R. J., Ochsenreither S., Dummer R., Kirkwood J. M., Joshua A. M., Sacco J. J., Shoushtari A. N., Orloff M., Piulats J. M., Milhem M., Salama A. K. S., Curti B., Demidov L., Gastaud L., Mauch C., Yushak M., Carvajal R. D., Hamid O., Abdullah S. E., Holland C., Goodall H., Piperno-Neumann S., IMCgp100–202 Investigators, Overall Survival Benefit with Tebentafusp in Metastatic Uveal Melanoma. N. Engl. J. Med. 385, 1196–1206 (2021). - PubMed
-
- Chandran S. S., Ma J., Klatt M. G., Dündar F., Bandlamudi C., Razavi P., Wen H. Y., Weigelt B., Zumbo P., Fu S. N., Banks L. B., Yi F., Vercher E., Etxeberria I., Bestman W. D., Da Cruz Paula A., Aricescu I. S., Drilon A., Betel D., Scheinberg D. A., Baker B. M., Klebanoff C. A., Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA. Nat. Med. 28, 946–957 (2022). - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
