This is a preprint.
Measuring renal cortical cell-specific mitochondrial metabolism
- PMID: 39651228
- PMCID: PMC11623503
- DOI: 10.1101/2024.11.24.622516
Measuring renal cortical cell-specific mitochondrial metabolism
Abstract
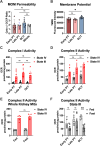
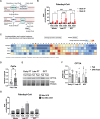
The metabolic health of the kidney is a primary determinant of the risk of progressive kidney disease. Our understanding of the metabolic processes that fuel kidney functions is limited by the kidney's structural and functional heterogeneity. As the kidney contains many different cell types, we sought to determine the intra-renal mitochondrial heterogeneity that contributes to cell-specific metabolism. To interrogate this, we utilized a recently developed mitochondrial tagging technique to isolate kidney cell-type specific mitochondria. Here, we investigate mitochondrial functional capacities and the metabolomes of the early and late proximal tubule (PT) and the distal convoluted tubule (DCT). The conditional MITO-Tag transgene was combined with Slc34a1-CreERT2, Ggt1-Cre, or Pvalb-Cre transgenes to generate mouse models capable of cell-specific isolation of hemagglutinin (HA)-tagged mitochondria from the early PT, late PT, or the DCT, respectively. Functional assays measuring mitochondrial respiratory and fatty acid oxidation (FAO) capacities and metabolomics were performed on anti-HA immunoprecipitated mitochondria from kidneys of ad libitum fed and 24-hour fasted male mice. The renal MITO-Tag models targeting the early PT, late PT, and DCT revealed differential mitochondrial respiratory and FAO capacities which dynamically changed during fasting conditions. The renal MITO-Tag model captured differential mitochondrial metabolism and functional capacities across the early PT, late PT, and DCT at baseline and in response to fasting.
Keywords: cellular metabolic heterogeneity; kidney; metabolism; mitochondria; tubular epithelium.
Figures



References
-
- Kappler L, Hoene M, Hu C, von Toerne C, Li J, Bleher D, Hoffmann C, Böhm A, Kollipara L, Zischka H, Königsrainer A, Häring HU, Peter A, Xu G, Sickmann A, Hauck SM, Weigert C, and Lehmann R. Linking bioenergetic function of mitochondria to tissue-specific molecular fingerprints. Am J Physiol Endocrinol Metab 317:E374–E387, 2019. 10.1152/ajpendo.00088.2019 - DOI - PubMed
-
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, and Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123, 2008. 10.1016/j.cell.2008.06.016 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
