Mode of Action and Mechanisms of Resistance to the Unusual Polyglycosylated Thiopeptide Antibiotic Persiathiacin A
- PMID: 39651842
- PMCID: PMC11731312
- DOI: 10.1021/acsinfecdis.4c00503
Mode of Action and Mechanisms of Resistance to the Unusual Polyglycosylated Thiopeptide Antibiotic Persiathiacin A
Abstract
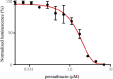
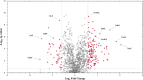
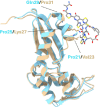
Persiathiacin A is a novel thiopeptide antibiotic produced by Actinokineospora species UTMC 2448. It has potent activity against methicillin-resistant Staphylococcus aureus (MRSA) and Mycobacterium tuberculosis. Thiopeptides, including persiathiacin A, exhibit antibacterial activity by inhibiting protein synthesis. In this study, we characterize the mechanism of action of persiathiacin A and investigate how resistance to this antibiotic can emerge. In vitro assays revealed that persiathiacin A inhibits translation elongation, leading to ribosome stalling. Genetic analysis of resistant Bacillus subtilis mutants identified mutations primarily in the rplK gene encoding ribosomal protein L11, which is the binding site for other 26-membered macrocycle-containing thiopeptides. The resistant mutants showed growth impairment and an increased lag time, even in the absence of persiathiacin. Comparative proteomic analysis of a resistant mutant versus the parental strain revealed multiple changes, indicative of negative effects on protein synthesis. Thus, although persiathiacin-resistant mutants can arise readily by the loss of L11 function, it is likely that such mutants would be severely compromised in pathogenesis. Furthermore, bioinformatics analysis identified differences in the key amino acids within the thiopeptide-binding region of L11 in the persiathiacin producer. These probably prevent the antibiotic from associating with its target, providing a mechanism for self-resistance.
Keywords: RiPP; Ribosomal protein L11; Self-resistance; Translation inhibition.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K.-D.; Fischbach M. A.; Garavelli J. S.; Göransson U.; Gruber C. W.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Mitchell D. A.; Moll G. N.; Moore B. S.; Müller R.; Nair S. K.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J. T.; Rebuffat S.; Ross R. P.; Sahl H.-G.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Süssmuth R. D.; Tagg J. R.; Tang G.-L.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30 (1), 108–160. 10.1039/C2NP20085F. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

