A STAG2-PAXIP1/PAGR1 axis suppresses lung tumorigenesis
- PMID: 39652422
- PMCID: PMC11627241
- DOI: 10.1084/jem.20240765
A STAG2-PAXIP1/PAGR1 axis suppresses lung tumorigenesis
Abstract
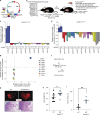
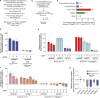
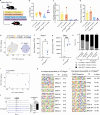
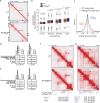
The cohesin complex is a critical regulator of gene expression. STAG2 is the most frequently mutated cohesin subunit across several cancer types and is a key tumor suppressor in lung cancer. Here, we coupled somatic CRISPR-Cas9 genome editing and tumor barcoding with an autochthonous oncogenic KRAS-driven lung cancer model and showed that STAG2 is uniquely tumor-suppressive among all core and auxiliary cohesin components. The heterodimeric complex components PAXIP1 and PAGR1 have highly correlated effects with STAG2 in human lung cancer cell lines, are tumor suppressors in vivo, and are epistatic to STAG2 in oncogenic KRAS-driven lung tumorigenesis in vivo. STAG2 inactivation elicits changes in gene expression, chromatin accessibility, and 3D genome conformation that impact the cancer cell state. Gene expression and chromatin accessibility similarities between STAG2- and PAXIP1-deficient neoplastic cells further relate STAG2-cohesin to PAXIP1/PAGR1. These findings reveal a STAG2-PAXIP1/PAGR1 tumor-suppressive axis and uncover novel PAXIP1-dependent and PAXIP1-independent STAG2-cohesin-mediated mechanisms of lung tumor suppression.
© 2024 Ashkin et al.
Conflict of interest statement
Disclosures: H.Y. Chang is a co-founder of Accent Therapeutics, Boundless Bio, Cartography Biosciences, and Orbital Therapeutics, and an advisor of 10x Genomics, Arsenal Bio, Chroma Medicine, Exai Bio, and Spring Discovery. M.M. Winslow and D.A. Petrov reported grants from the NIH (R01-CA231253, R01-CA234349, and P30-CA124435) during the conduct of the study and are founders and equity holders in Guide Oncology Inc. No other disclosures were reported.
Figures








Update of
-
A STAG2-PAXIP1/PAGR1 axis suppresses lung tumorigenesis.bioRxiv [Preprint]. 2024 Sep 19:2024.09.14.613043. doi: 10.1101/2024.09.14.613043. bioRxiv. 2024. Update in: J Exp Med. 2025 Jan 6;222(1):e20240765. doi: 10.1084/jem.20240765. PMID: 39345539 Free PMC article. Updated. Preprint.
References
-
- AACR Project GENIE Consortium . 2017. AACR Project GENIE: Powering precision medicine through an international consortium. Cancer Discov. 7:818–831. 10.1158/2159-8290.CD-17-0151 - DOI - PMC - PubMed
-
- Arruda, N.L., Carico Z.M., Justice M., Liu Y.F., Zhou J., Stefan H.C., Dowen J.M., Lee J.T., and Joyce E.F.. 2020. Distinct and overlapping roles of STAG1 and STAG2 in cohesin localization and gene expression in embryonic stem cells. Epigenetics Chromatin. 13:32. 10.1186/s13072-020-00353-9 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- T32 CA009302/CA/NCI NIH HHS/United States
- F99CA284289/CA/NCI NIH HHS/United States
- F99 CA274692/CA/NCI NIH HHS/United States
- 28FT-0019/Tobacco-Related Disease Research Program
- Stanford University
- K00 CA245784/CA/NCI NIH HHS/United States
- DGE-2146755/National Science Foundation Graduate Research Fellowship Program
- MFE-176568/Canadian Institute of Health Research
- R01 CA234349/CA/NCI NIH HHS/United States
- F99 CA284289/CA/NCI NIH HHS/United States
- P30 CA124435/CA/NCI NIH HHS/United States
- GT14928/HHMI/Howard Hughes Medical Institute/United States
- R01 CA231253/CA/NCI NIH HHS/United States
- R01-CA231253/NH/NIH HHS/United States
- PF-21-112-01-MM/American Cancer Society
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

