Targeting Arachidonic Acid Metabolism Enhances Immunotherapy Efficacy in ARID1A-Deficient Colorectal Cancer
- PMID: 39652583
- PMCID: PMC11873721
- DOI: 10.1158/0008-5472.CAN-24-1611
Targeting Arachidonic Acid Metabolism Enhances Immunotherapy Efficacy in ARID1A-Deficient Colorectal Cancer
Abstract
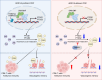
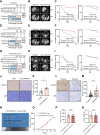
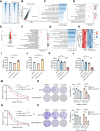
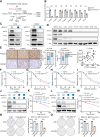
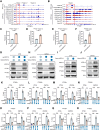
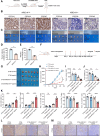
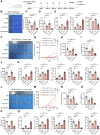
AT-rich interactive domain-containing protein 1A (ARID1A), a core constituent of the switch/sucrose nonfermentable (SWI/SNF) complex, is mutated in approximately 10% of colorectal cancers. Whereas ARID1A deficiency corresponds to heightened immune activity in colorectal cancer, immune checkpoint inhibitors (ICI) have shown limited efficacy in these tumors. The discovery of targetable vulnerabilities associated with ARID1A deficiency in colorectal cancer could expand treatment options for patients. In this study, we demonstrated that arachidonic acid (AA) metabolism inhibitors synergize with ICIs in ARID1A-deficient colorectal cancer by enhancing the activity of CD8+ T cells and inhibiting vasculogenic mimicry. Epigenetic analysis using ATAC-seq and ChIP-qPCR revealed that the lack of ARID1A results in reduced levels of PTGS1 and PTGS2, the key enzymes that control the AA pathway. Low PTGS1 and PTGS2 expression generated a reliance on the remaining functionality of the AA pathway in ARID1A-deficient cells. The AA pathway inhibitor aspirin selectively inhibited the growth of ARID1A-deficient colorectal cancer, and aspirin sensitized tumors lacking ARID1A to immunotherapy. Together, these findings suggest that blocking AA metabolism can enhance immune responses against tumors by activating CD8+ T cells and inhibiting vasculogenic mimicry, which synergizes with ICIs to improve treatment of ARID1A-deficient colorectal cancer. Significance: The arachidonic acid pathway is a metabolic vulnerability in ARID1A-deficient colorectal cancer that can be targeted with aspirin to suppress tumor growth and enhance sensitivity to immunotherapy, providing a promising therapeutic strategy.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
No disclosures were reported.
Figures

References
-
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009;78:273–304. - PubMed
-
- Bosse T, ter Haar NT, Seeber LM, v Diest PJ, Hes FJ, Vasen HF, et al. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod Pathol 2013;26:1525–35. - PubMed
MeSH terms
Substances
Grants and funding
- 2022ZX06C01/Key Research and Development Program of Heilongjiang ()
- JD2023SJ40/Key Research and Development Program of Heilongjiang ()
- YQ2022H017/Natural Science Foundation of Heilongjiang Province (Heilongjiang Natural Science Foundation)
- U22A20330/National Natural Science Foundation of China (NSFC)
- 82373372/National Natural Science Foundation of China (NSFC)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

