Comparative study of two Rift Valley fever virus field strains originating from Mauritania
- PMID: 39652604
- PMCID: PMC11658707
- DOI: 10.1371/journal.pntd.0012728
Comparative study of two Rift Valley fever virus field strains originating from Mauritania
Abstract
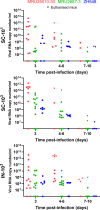
Rift Valley fever (RVF) is one of the major viral arthropod-borne diseases in Africa. In recent decades, RVF virus (RVFV), the causative agent of RVF, has been responsible for multiple outbreaks in West Africa with important consequences on human and animal health. In particular, an outbreak occurred in 2010 after heavy rains in the desertic region of Adrar, Mauritania. It was characterized by the appearance of severe clinical signs among dromedary camels. Another one occurred in 2013-2014 across Senegal and the southern part of Mauritania. In this study, we characterized two RVFV field strains isolated during these two outbreaks. The first strain, MRU25010-30, was isolated from a camel (2010) while the second, MRU2687-3, was isolated from a goat (2013). By deep-sequencing and rapid amplification of cDNA-ends by polymerase chain reaction, we successfully sequenced the complete genome of these two RVFV strains as well as the reference laboratory strain ZH548. Phylogenetic analysis showed that the two field viruses belong to two different RVFV genetic lineages. Moreover, we showed that MRU25010-30 replicates more efficiently in various in vitro cell culture models than MRU2687-3 and ZH548. In vivo, MRU25010-30 caused rapid death of BALB/c mice and proved to be more virulent than MRU2687-3, regardless of the route of inoculation (subcutaneous or intranasal). The virulence of MRU25010-30 is associated with a high viral load in the liver and serum of infected mice, while the death of mice infected with MRU2687-3 and ZH548 correlated with a high viral load in the brain. Altogether, the data presented in this study provide new avenues to unveil the molecular viral determinants that modulate RVFV virulence and replication capacity.
Copyright: © 2024 Chabert et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Daubney R, Hudson JR, Garnham PC. Enzootic hepatitis or Rift Valley fever. An undescribed virus disease of sheep cattle and man from East Africa. J Pathol Bacteriol. 1931;34: 545–579. doi: 10.1002/path.1700340418 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

