Cbf11 and Mga2 function together to activate transcription of lipid metabolism genes and promote mitotic fidelity in fission yeast
- PMID: 39652606
- PMCID: PMC11658701
- DOI: 10.1371/journal.pgen.1011509
Cbf11 and Mga2 function together to activate transcription of lipid metabolism genes and promote mitotic fidelity in fission yeast
Abstract
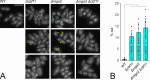
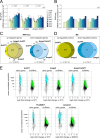
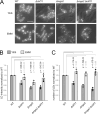
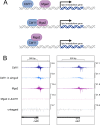
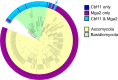
Within a eukaryotic cell, both lipid homeostasis and faithful cell cycle progression are meticulously orchestrated. The fission yeast Schizosaccharomyces pombe provides a powerful platform to study the intricate regulatory mechanisms governing these fundamental processes. In S. pombe, the Cbf11 and Mga2 proteins are transcriptional activators of non-sterol lipid metabolism genes, with Cbf11 also known as a cell cycle regulator. Despite sharing a common set of target genes, little was known about their functional relationship. This study reveals that Cbf11 and Mga2 function together in the same regulatory pathway, critical for both lipid metabolism and mitotic fidelity. Deletion of either gene results in a similar array of defects, including slow growth, dysregulated lipid homeostasis, impaired cell cycle progression (cut phenotype), abnormal cell morphology, perturbed transcriptomic and proteomic profiles, and compromised response to the stressors camptothecin and thiabendazole. Remarkably, the double deletion mutant does not exhibit a more severe phenotype compared to the single mutants. In addition, ChIP-nexus analysis reveals that both Cbf11 and Mga2 bind to nearly identical positions within the promoter regions of target genes. Interestingly, Mga2 binding appears to be dependent on the presence of Cbf11 and Cbf11 likely acts as a tether to DNA, while Mga2 is needed to activate the target genes. In addition, the study explores the distribution of Cbf11 and Mga2 homologs across fungi. The presence of both Cbf11 and Mga2 homologs in Basidiomycota contrasts with Ascomycota, which mostly lack Cbf11 but retain Mga2. This suggests an evolutionary rewiring of the regulatory circuitry governing lipid metabolism and mitotic fidelity. In conclusion, this study offers compelling support for Cbf11 and Mga2 functioning jointly to regulate lipid metabolism and mitotic fidelity in fission yeast.
Copyright: © 2024 Marešová et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
CSL protein regulates transcription of genes required to prevent catastrophic mitosis in fission yeast.Cell Cycle. 2016 Nov 16;15(22):3082-3093. doi: 10.1080/15384101.2016.1235100. Epub 2016 Sep 29. Cell Cycle. 2016. PMID: 27687771 Free PMC article.
-
Mga2 Transcription Factor Regulates an Oxygen-responsive Lipid Homeostasis Pathway in Fission Yeast.J Biol Chem. 2016 Jun 3;291(23):12171-83. doi: 10.1074/jbc.M116.723650. Epub 2016 Apr 6. J Biol Chem. 2016. PMID: 27053105 Free PMC article.
-
Coordinate Regulation of Yeast Sterol Regulatory Element-binding Protein (SREBP) and Mga2 Transcription Factors.J Biol Chem. 2017 Mar 31;292(13):5311-5324. doi: 10.1074/jbc.M117.778209. Epub 2017 Feb 15. J Biol Chem. 2017. PMID: 28202541 Free PMC article.
-
Genes affecting the extension of chronological lifespan in Schizosaccharomyces pombe (fission yeast).Mol Microbiol. 2021 Apr;115(4):623-642. doi: 10.1111/mmi.14627. Epub 2020 Nov 3. Mol Microbiol. 2021. PMID: 33064911 Free PMC article. Review.
-
The phenomenon of lipid metabolism "cut" mutants.Yeast. 2018 Dec;35(12):631-637. doi: 10.1002/yea.3358. Epub 2018 Oct 29. Yeast. 2018. PMID: 30278108 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

