Fatal Lassa fever in cynomolgus monkeys is associated with systemic viral dissemination and inflammation
- PMID: 39652618
- PMCID: PMC11658700
- DOI: 10.1371/journal.ppat.1012768
Fatal Lassa fever in cynomolgus monkeys is associated with systemic viral dissemination and inflammation
Abstract
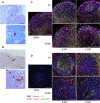
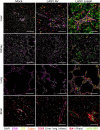
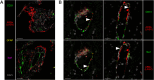
The pathogenesis of Lassa fever has not yet been fully deciphered, particularly as concerns the mechanisms determining whether acute infection is controlled or leads to catastrophic illness and death. Using a cynomolgus monkey model of Lassa virus (LASV) infection reproducing the different outcomes of the disease, we performed histological and transcriptomic studies to investigate the dynamics of LASV infection and the immune mechanisms associated with survival or death. Lymphoid organs are an early major reservoir for replicating virus during Lassa fever, with LASV entering through the cortical sinus of draining lymph nodes regardless of disease outcome. However, subsequent viral tropism varies considerably with disease severity, with viral dissemination limited almost entirely to lymphoid organs and immune cells during nonfatal Lassa fever. By contrast, the systemic dissemination of LASV to all organs and diverse cell types, leading to infiltrations with macrophages and neutrophils and an excessive inflammatory response, is associated with a fatal outcome. These results provide new insight into early viral dynamics and the host response to LASV infection according to disease outcome.
Copyright: © 2024 Hortion et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Buchmeier MJ, de la Torre J-C, Peters CJ. Arenaviridae. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, et al., editors. Fields virology. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. p. 1283–303.
MeSH terms
LinkOut - more resources
Full Text Sources

