Butyrate-producing Faecalibacterium prausnitzii suppresses natural killer/T-cell lymphoma by dampening the JAK-STAT pathway
- PMID: 39653411
- PMCID: PMC12013593
- DOI: 10.1136/gutjnl-2024-333530
Butyrate-producing Faecalibacterium prausnitzii suppresses natural killer/T-cell lymphoma by dampening the JAK-STAT pathway
Abstract
Background: Natural killer/T-cell lymphoma (NKTCL) is a highly aggressive malignancy with a dismal prognosis, and gaps remain in understanding the determinants influencing disease outcomes.
Objective: To characterise the gut microbiota feature and identify potential probiotics that could ameliorate the development of NKTCL.
Design: This cross-sectional study employed shotgun metagenomic sequencing to profile the gut microbiota in two Chinese NKTCL cohorts, with validation conducted in an independent Korean cohort. Univariable and multivariable Cox proportional hazards analyses were applied to assess associations between identified marker species and patient outcomes. Tumour-suppressing effects were investigated using comprehensive in vivo and in vitro models. In addition, metabolomics, RNA sequencing, chromatin immunoprecipitation sequencing, Western blot analysis, immunohistochemistry and lentiviral-mediated gene knockdown system were used to elucidate the underlying mechanisms.
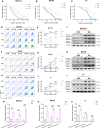
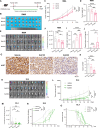
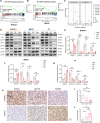
Results: We first unveiled significant gut microbiota dysbiosis in NKTCL patients, prominently marked by a notable reduction in Faecalibacterium prausnitzii which correlated strongly with shorter survival among patients. Subsequently, we substantiated the antitumour properties of F. prausnitzii in NKTCL mouse models. Furthermore, F. prausnitzii culture supernatant demonstrated significant efficacy in inhibiting NKTCL cell growth. Metabolomics analysis revealed butyrate as a critical metabolite underlying these tumour-suppressing effects, validated in three human NKTCL cell lines and multiple tumour-bearing mouse models. Mechanistically, butyrate suppressed the activation of Janus kinase-signal transducer and activator of transcription pathway through enhancing histone acetylation, promoting the expression of suppressor of cytokine signalling 1.
Conclusion: These findings uncover a distinctive gut microbiota profile in NKTCL and provide a novel perspective on leveraging the therapeutic potential of F. prausnitzii to ameliorate this malignancy.
Keywords: BUTYRATE; INTESTINAL MICROBIOLOGY; LYMPHOMA; PROBIOTICS.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures




References
-
- Lim SH, Hong JY, Lim ST, et al. Beyond first-line non-anthracycline-based chemotherapy for extranodal NK/T-cell lymphoma: clinical outcome and current perspectives on salvage therapy for patients after first relapse and progression of disease. Ann Oncol. 2017;28:2199–205. doi: 10.1093/annonc/mdx316. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
