DNA barcoded peptide-MHC multimers to measure and monitor minor histocompatibility antigen-specific T cells after allogeneic stem cell transplantation
- PMID: 39653555
- PMCID: PMC11629015
- DOI: 10.1136/jitc-2024-009564
DNA barcoded peptide-MHC multimers to measure and monitor minor histocompatibility antigen-specific T cells after allogeneic stem cell transplantation
Abstract
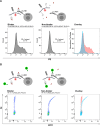
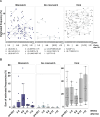
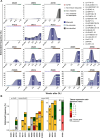
Allogeneic stem cell transplantation (alloSCT) provides a curative treatment option for hematological malignancies. After HLA-matched alloSCT, donor-derived T cells recognize minor histocompatibility antigens (MiHAs), which are polymorphic peptides presented by HLA on patient cells. MiHAs are absent on donor cells due to genetic differences between patient and donor. T cells targeting broadly expressed MiHAs induce graft-versus-leukemia (GvL) reactivity as well as graft-versus-host disease (GvHD), while T cells for MiHAs with restricted or preferential expression on hematopoietic or non-hematopoietic cells may skew responses toward GvL or GvHD, respectively. Besides tissue expression, overall strength of GvL and GvHD is also determined by T-cell frequencies against MiHAs.Here, we explored the use of DNA barcode-labeled peptide-MHC multimers to detect and monitor antigen-specific T cells for the recently expanded repertoire of HLA-I-restricted MiHAs. In 16 patients who experienced an immune response after donor lymphocyte infusion, variable T-cell frequencies up to 30.5% of CD8+ T cells were measured for 49 MiHAs. High T-cell frequencies above 1% were measured in 12 patients for 19 MiHAs, with the majority directed against mismatched MiHAs, typically 6-8 weeks after donor lymphocyte infusion and at the onset of GvHD. The 12 patients included 9 of 10 patients with severe GvHD, 2 of 3 patients with limited GvHD and 1 of 3 patients without GvHD.In conclusion, we demonstrated that barcoded peptide-MHC multimers reliably detect and allow monitoring for MiHA-specific T cells during treatment to investigate the kinetics of immune responses and their impact on development of GvL and GvHD after HLA-matched alloSCT.
Keywords: Graft versus host disease - GVHD; Graft versus leukemia; Hematologic Malignancies; T cell; Transplant.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
-
- van der Zouwen B, Koster EAS, von dem Borne PA, et al. Feasibility, safety, and efficacy of early prophylactic donor lymphocyte infusion after T cell-depleted allogeneic stem cell transplantation in acute leukemia patients. Ann Hematol. 2023;102:1203–13. doi: 10.1007/s00277-023-05145-1. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
