Esophageal ILC2s mediate abnormal epithelial remodeling in eosinophilic esophagitis via Areg-EGFR signaling
- PMID: 39653767
- PMCID: PMC11685411
- DOI: 10.1038/s41423-024-01242-x
Esophageal ILC2s mediate abnormal epithelial remodeling in eosinophilic esophagitis via Areg-EGFR signaling
Abstract
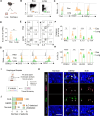
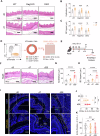
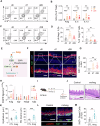
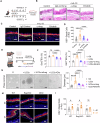
Eosinophilic esophagitis (EoE) is a chronic allergic disorder characterized by eosinophilia and epithelial thickening, resulting in dysphagia. While emerging evidence implicates increased frequencies of group 2 innate lymphoid cells (ILC2s) and increased interleukin (IL)-33 expression in EoE pathogenesis, the precise mechanisms remain unclear. In this study, we investigated the role of ILC2s in EoE pathogenesis. We observed an abundance of KLRG1+ ILC2s in the esophagi of healthy mice, with their numbers significantly increasing in murine EoE models and humans. Using a murine EoE model, we demonstrated the recapitulation of EoE-associated features, including basal-cell hyperproliferation, epithelial thickening, and eosinophilia. Notably, these characteristics are absent in ILC-deficient mice, whereas mice lacking IL-5 or eosinophils display epithelial defects, highlighting the pivotal role of ILC2s in EoE pathogenesis. Further investigations revealed increased amphiregulin (Areg) production by esophageal ILC2s in mice. The administration of Areg induced epithelial defects similar to those observed in EoE. Mechanistic studies using human esophageal cell lines revealed Areg-induced phosphorylation of epidermal growth factor receptor (EGFR). Significatntly, treatment with anti-Areg agents and EGFR inhibitors effectively attenuated EoE development, highlighting the therapeutic potential of targeting the Areg-EGFR axis.
Keywords: Allergic disorder; Amphiregulin (Areg); Eosinophilic esophagitis (EoE); Epidermal growth factor receptor (EGFR); Epidermal hyperplasia; Innate lymphoid cells (ILCs).
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Mulder DJ, Justinich CJ. Understanding eosinophilic esophagitis: the cellular and molecular mechanisms of an emerging disease. Mucosal Immunol. 2011;4:139–47. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

