Dissociable control of motivation and reinforcement by distinct ventral striatal dopamine receptors
- PMID: 39653808
- PMCID: PMC12065418
- DOI: 10.1038/s41593-024-01819-9
Dissociable control of motivation and reinforcement by distinct ventral striatal dopamine receptors
Abstract
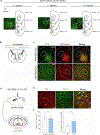
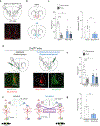
Dopamine (DA) release in striatal circuits, including the nucleus accumbens medial shell (mNAcSh), tracks separable features of reward like motivation and reinforcement. However, the cellular and circuit mechanisms by which DA receptors transform DA release into distinct constructs of reward remain unclear. Here we show that DA D3 receptor (D3R) signaling in the mNAcSh drives motivated behavior in mice by regulating local microcircuits. Furthermore, D3Rs coexpress with DA D1 receptors, which regulate reinforcement, but not motivation. Paralleling dissociable roles in reward function, we report nonoverlapping physiological actions of D3R and DA D1 receptor signaling in mNAcSh neurons. Our results establish a fundamental framework wherein DA signaling within the same nucleus accumbens cell type is physiologically compartmentalized via actions on distinct DA receptors. This structural and functional organization provides neurons in a limbic circuit with the unique ability to orchestrate dissociable aspects of reward-related behaviors relevant to the etiology of neuropsychiatric disorders.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures













Update of
-
Dissociable control of motivation and reinforcement by distinct ventral striatal dopamine receptors.bioRxiv [Preprint]. 2023 Jun 28:2023.06.27.546539. doi: 10.1101/2023.06.27.546539. bioRxiv. 2023. Update in: Nat Neurosci. 2025 Jan;28(1):105-121. doi: 10.1038/s41593-024-01819-9. PMID: 37425766 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 ES034037/ES/NIEHS NIH HHS/United States
- ZIA MH002970-04/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R01 DA061243/DA/NIDA NIH HHS/United States
- ZIA MH002970/ImNIH/Intramural NIH HHS/United States
- 29671/Brain and Behavior Research Foundation (Brain & Behavior Research Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

