Erucic acid utilization by Lactobacillus johnsonii N6.2
- PMID: 39654680
- PMCID: PMC11625735
- DOI: 10.3389/fmicb.2024.1476958
Erucic acid utilization by Lactobacillus johnsonii N6.2
Abstract
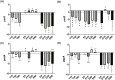
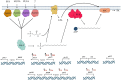
A multivariate nutritional analysis indicated that the consumption of erucic acid-rich food, a fatty acid (FA) found primarily in rapeseed and mustard oil, was positively correlated with higher counts of lactic acid bacteria (LAB). Furthermore, we showed Lactobacillus johnsonii N6.2, as well as other species of LAB tested from the former Lactobacillus genus, were able to efficiently use erucic acid (EA) as the source of FA. In this work, we identified significant changes induced in the FA profiles of L. johnsonii cultured with EA as the source of FA. We performed global transcriptomics to identify genes and pathways involved in EA utilization. It was found that L. johnsonii incorporates external fatty acids via a FakA/FakB and the plsX/plsY/plsC pathway for phosphatidic acid synthesis. It was found that cells grown in MRS with EA (MRS-E) significantly upregulated fakB2 and fakB4 when compared to cells grown in standard MRS with tween 80 as the source of FA. Additionally, in MRS-E, L. johnsonii N6.2 induced the expression of plsY2, plsC2 and plsC4 while the expression of pslX was constitutive during short term EA exposure. LC-MS analyses revealed that L. johnsonii N6.2 rapidly incorporates EA and synthesizes a variety of long chain fatty acids, including the health-relevant omega-9 monounsaturated fatty acids such as nervonic and gondoic acids.
Keywords: Lactobacillus johnsonii; erucic acid; long chain fatty acid; nervonic acid; probiotic.
Copyright © 2024 Thompson, Beliakoff, Garrett, Gonzalez and Lorca.
Conflict of interest statement
GL holds U.S. patent no. 9474773 and 9987313 on Lactobacillus johnsonii N6.2. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

