Epigenomic and clinical analyses of striatal DAT binding in healthy individuals reveal well-known loci of Parkinson's disease
- PMID: 39654757
- PMCID: PMC11625257
- DOI: 10.1016/j.heliyon.2024.e40618
Epigenomic and clinical analyses of striatal DAT binding in healthy individuals reveal well-known loci of Parkinson's disease
Abstract
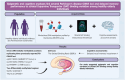
Background: Striatal dopamine transporter (DAT) binding is a sensitive and specific endophenotype for detecting dopaminergic deficits across Parkinson's disease (PD) spectrum. Molecular and clinical signatures of PD in asymptomatic phases help understand the earliest pathophysiological mechanisms underlying the disease. We aimed to investigate whether blood epigenetic markers are associated with inter-individual variation of striatal DAT binding among healthy elderly individuals. We also investigated whether this potential inter-individual variation can manifest as dysfunction of particular cognitive domains. Omics studies conducted on endophenotypes of PD among healthy asymptomatic individuals can provide invaluable insights into early detection, disease mechanisms, and potential therapeutic targets for PD.
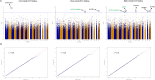
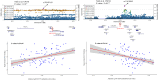
Method: We conducted a blood epigenome-wide association study of striatal DAT binding on 96 healthy individuals using the Illumina EPIC array. For functional annotation of our top results, we employed the enhancer-gene mapping strategy using a midbrain single-nucleus multimodal dataset. Finally, we conducted several investigative regression analyses on several neuropsychological tests across five cognitive domains to assess their association with striatal DAT binding among 250 healthy subjects.
Results: We identified seven suggestive (P-value<10-5) CpG probes. Specifically, three probes were colocalized with three risk loci previously identified in PD's largest Genome-Wide Association Study (GWAS). UCN5A and APOE loci were identified as suggestive DMRs associated with striatal DAT binding. Functional analyses prioritized the FDFT1 gene as the potential target gene in the previously reported CTSB GWAS locus. We also showed that delayed recall memory impairment was correlated with reduced striatal DAT binding, irrespective of age.
Conclusion: Our study suggested epigenetic and cognitive signatures of striatal DAT binding among healthy individuals, providing valuable insights for future experimental and clinical studies of early PD.
Keywords: Cognitive domains; DAT scan; Epigenome-wide association study; Healthy individuals; Parkinson's disease.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

