Evaluation of intranasal TLR2/6 agonist INNA-051: safety, tolerability and proof of pharmacology
- PMID: 39655168
- PMCID: PMC11626610
- DOI: 10.1183/23120541.00199-2024
Evaluation of intranasal TLR2/6 agonist INNA-051: safety, tolerability and proof of pharmacology
Abstract
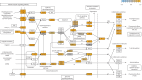
Background: Local priming of the innate immune system with a Toll-like receptor (TLR)2/6 agonist may reduce morbidity and mortality associated with viral respiratory tract infections, particularly for the elderly and those with chronic diseases. The objectives of the present study were to understand the potential of prophylactic treatment with a TLR2/6 agonist as an enhancer of innate immunity pathways leading to accelerated respiratory virus clearance from the upper airways.
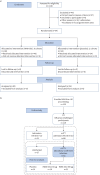
Methods: Two randomised, double-blind, placebo-controlled clinical trials were conducted in healthy adult participants. The first dose-escalation study assessed safety, tolerability and mechanistic biomarkers following single and repeated intranasal administrations of INNA-051. The second was an influenza A viral challenge study assessing the impact of treatment on host defence biomarkers and viral load.
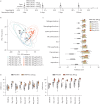
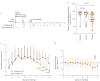
Results: INNA-051 was well tolerated in both studies, with no dose-limiting toxicities identified. Mechanistic biomarkers assessed in both studies demonstrated the expected engagement of pharmacology, including innate immune pathways. There were lower than anticipated rates of infection. Post hoc analysis conducted in laboratory-confirmed infected participants with low or no antibody titre against the challenge virus showed INNA-051 treatment led to a significantly shorter duration of infection and increased expression of genes and pathways associated with host defence responses against influenza.
Conclusions: The safety and pharmacology profile of INNA-051 confirms preclinical studies. INNA-051 increased expression of genes and pathways associated with host defence responses against influenza and was associated with a shorter duration of infection. These studies support further clinical assessment in the context of natural viral respiratory tract infections in individuals at increased risk of severe illness.
Copyright ©The authors 2024.
Conflict of interest statement
Conflict of interest: F.A. Mercuri, C. Demaison and G. McLachlan are employees of ENA Respiratory Pty Ltd, and receive an annual salary and other benefits. In addition, they have shares and share options and are named inventors on numerous granted or pending patent applications controlled by ENA. Conflict of interest: H.A. McQuilten receives payments as a contract researcher from ENA Respiratory Pty Ltd. S. White, R. Tal-Singer, B.E. Miller and N. Kruger receive payments as paid consultants for ENA Respiratory Pty Ltd, and as part of their compensation have been awarded participation in the company share option plan. R. Tal-Singer is a retiree and shareholder of GSK, and reports personal fees from AstraZeneca, Roche, Vocalis Health, Teva, ImmunoMet, Renovion, Samay Health, GSK and ItayAndBeyond. Conflict of interest: S. Mynhardt, N.P. West, P. Zhang, C. Lemech and R. Hari receive payments to their institutions for carrying out research.
Figures


References
LinkOut - more resources
Full Text Sources
