Discovery of Cryptic Natural Products Using High-Throughput Elicitor Screening on Agar Media
- PMID: 39655417
- PMCID: PMC12303134
- DOI: 10.1021/acs.biochem.4c00659
Discovery of Cryptic Natural Products Using High-Throughput Elicitor Screening on Agar Media
Abstract
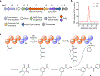
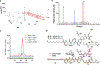
It is now well-established that microbial genomes carry sparingly expressed biosynthetic gene clusters (BGCs) that need to be induced in order to characterize their products. To do so, we herein subjected two well-known producers, Burkholderia plantarii and Burkholderia gladioli, to high-throughput elicitor screening (HiTES), an emerging approach for accessing the products of these "cryptic" BGCs. Both organisms have previously been examined extensively in liquid cultures. We therefore applied HiTES on agar and found several novel natural products that are only produced in this format and not in liquid cultures. Most notably we found two metabolites, termed burkethyl A and B, that contain an unusual m-ethylbenzoyl group and for which we identified the cognate BGC using bioinformatic and genetic studies. Our results indicate that agar-based HiTES is a promising approach for natural product discovery and are in line with the notion that even "drained" strains remain sources of new metabolites as long as alternative approaches are employed.
Conflict of interest statement
Notes
The authors declare the following competing financial interest(s): M.R.S. is co-founder of Cryptyx Bioscience and consultant to Merck Co. These entities played no role in the current study.
Figures

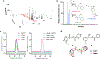
References
-
- Newman DJ; Cragg GM Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. - PubMed
-
- Clardy J; Walsh C Lessons from natural molecules. Nature 2004, 432, 829–837. - PubMed
-
- Rutledge PJ; Challis GL Discovery of Microbial Natural Products by Activation of Silent Biosynthetic Gene Clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

