Enzymatic Reaction Network-Driven Polymerization-Induced Transient Coacervation
- PMID: 39655501
- PMCID: PMC11891636
- DOI: 10.1002/anie.202421620
Enzymatic Reaction Network-Driven Polymerization-Induced Transient Coacervation
Abstract
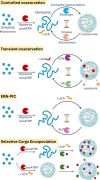
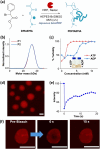
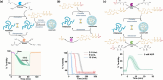
A living cell has a highly complex microenvironment whereas numerous enzyme-driven processes are active at once. These procedures are incredibly accurate and efficient, although comparable control has not yet been established in vitro. Here, we design an enzymatic reaction network (ERN) that combines antagonistic and orthogonal enzymatic networks to produce adjustable dynamics of ATP-fueled transient coacervation. Using horseradish peroxidase (HRP)-mediated Biocatalytic Atom Transfer Radical Polymerization (BioATRP), we synthesized poly(dimethylaminoethyl methacrylate), which subsequently formed coacervates with ATP. We rationally explored enzymatic control over coacervation and dissolution, using orthogonal and antagonistic enzyme pairs viz., alkaline phosphatase, Creatine phosphokinase, hexokinase, esterase, and urease. ATP-fuelled coacervates also demonstrate the enzymatic catalysis to prove its potential to be exploited as a cellular microreactor. Additionally, we developed ERN-polymerization-induced transient coacervation (ERN-PIC), with complete control over the system, polymerization, coacervation, and dissolution. Notably, the coacervation process itself determines functional properties, as seen in selective cargo uptake. The strategy offers cutting-edge biomimetic applications, and insights into cellular compartmentalization by bridging the gap between synthetic and biological systems. The development of temporally programmed coacervation is promising for the spatial arrangement of multienzyme cascades, and offers novel ideas on the architecture of artificial cells.
Keywords: ATP; BioATRP; Coacervates; Enzymatic Reaction Network; Liquid-liquid phase separation.
© 2024 The Author(s). Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Huber M. C., Schreiber A., von Olshausen P., Varga B. R., Kretz O., Joch B., Barnert S., Schubert R., Eimer S., Kele P., Schiller S. M., Nat. Mater. 2015, 14, 125–132. - PubMed
-
- Roodbeen R., van Hest J. C. M., BioEssays 2009, 31, 1299–1308. - PubMed
-
- Lin Z., Beneyton T., Baret J.-C., Martin N., Small Methods 2023, 7, 2300496. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

