New multilocus sequence typing scheme for Enterococcus faecium reveals sequential outbreaks of vancomycin-resistant E. faecium ST1162 and ST610 in a Japanese tertiary medical center
- PMID: 39656011
- PMCID: PMC11705941
- DOI: 10.1128/spectrum.02131-24
New multilocus sequence typing scheme for Enterococcus faecium reveals sequential outbreaks of vancomycin-resistant E. faecium ST1162 and ST610 in a Japanese tertiary medical center
Abstract
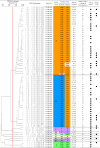
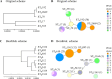
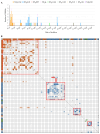
Vancomycin-resistant Enterococcus faecium (VREfm) is a major nosocomial pathogen, and molecular epidemiological tools are crucial for controlling its spread. Pulsed-field gel electrophoresis (PFGE) is still used in clinical laboratories despite the increased accessibility of whole-genome sequencing (WGS). As PFGE equipment is no longer commercially available, clinical laboratories need alternative tools. Highly standardized multilocus sequence typing (MLST) is one option. However, the original MLST scheme for E. faecium, designed in 2002, showed inconsistencies with WGS-based typing. Therefore, the new Bezdíček MLST scheme, which offers more accurate genetic similarity based on genome-wide data, has recently been proposed. To clarify its clinical utility in analyzing nosocomial VREfm transmission, we compared both MLST schemes with PFGE using 68 VREfm isolates collected during an outbreak at a Japanese tertiary medical center in 2019. PFGE analysis identified nine clusters among the 68 strains, including two predominant clusters. The original scheme identified five sequence types (STOs), of which 82.4% (56/68) were STO192. The Bezdíček scheme identified nine sequence types (STBs), subdividing the original STO192 into STB1162 (30/56), STB610 (25/56), and STB895 (1/56). Simpson's index of diversity values were 0.635, 0.317, and 0.648 for PFGE, the original scheme, and the Bezdíček scheme, respectively. Combining the Bezdíček scheme with admission records provided clearer outbreak visualization, indicating that two distinct STBs independently caused sequential outbreaks. With high discriminatory power comparable with PFGE and global availability, the Bezdíček scheme is a practical and valuable tool for controlling nosocomial VREfm infections in clinical laboratories.IMPORTANCEIn areas where vancomycin-resistant Enterococcus faecium is common, hospital-acquired infections pose a considerable threat to patients' lives owing to treatment difficulties. Although whole-genome sequencing-based typing has logically become the new reference standard and its accessibility is growing, many clinical laboratories still lack the fundamental resources to exploit its full potential. Limited availability of the traditional pulsed-field gel electrophoresis test in clinical settings has necessitated the use of alternative tools such as Bezdíček multilocus sequence typing. This study tested the clinical utility of the Bezdíček scheme by comparing it with pulsed-field gel electrophoresis. Designed using Czech isolates, this scheme showed comparable discriminatory powers with the traditional method for geographically distinct Japanese isolates and clearly visualized outbreaks. These findings suggest that the Bezdíček scheme is a potential alternative to pulsed-field gel electrophoresis for identifying hospital transmission of vancomycin-resistant Enterococcus faecium in clinical laboratories.
Keywords: Enterococcus faecium; multilocus sequence typing; outbreak; pulsed-field gel electrophoresis; vancomycin-resistant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011–2014. Infect Control Hosp Epidemiol 37:1288–1301. doi: 10.1017/ice.2016.174 - DOI - PMC - PubMed
-
- van Belkum A, van Leeuwen W, Kaufmann ME, Cookson B, Forey F, Etienne J, Goering R, Tenover F, Steward C, O’Brien F, Grubb W, Tassios P, Legakis N, Morvan A, El Solh N, de Ryck R, Struelens M, Salmenlinna S, Vuopio-Varkila J, Kooistra M, Talens A, Witte W, Verbrugh H. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J Clin Microbiol 36:1653–1659. doi: 10.1128/JCM.36.6.1653-1659.1998 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

