Developmental and Adult Striatal Patterning of Nociceptin Ligand Marks Striosomal Population With Direct Dopamine Projections
- PMID: 39656141
- PMCID: PMC11629859
- DOI: 10.1002/cne.70003
Developmental and Adult Striatal Patterning of Nociceptin Ligand Marks Striosomal Population With Direct Dopamine Projections
Abstract
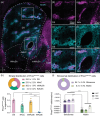
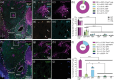
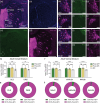
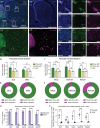
Circuit influences on the midbrain dopamine system are crucial to adaptive behavior and cognition. Recent developments in the study of neuropeptide systems have enabled high-resolution investigations of the intersection of neuromodulatory signals with basal ganglia circuitry, identifying the nociceptin/orphanin FQ (N/OFQ) endogenous opioid peptide system as a prospective regulator of striatal dopamine signaling. Using a prepronociceptin-Cre reporter mouse line, we characterized highly selective striosomal patterning of Pnoc mRNA expression in mouse dorsal striatum, reflecting the early developmental expression of Pnoc. In the ventral striatum, Pnoc expression in the nucleus accumbens core was grouped in clusters akin to the distribution found in striosomes. We found that PnoctdTomato reporter cells largely comprise a population of dopamine receptor D1 (Drd1) expressing medium spiny projection neurons localized in dorsal striosomes, known to be unique among striatal projection neurons for their direct innervation of midbrain dopamine neurons. These findings provide a new understanding of the intersection of the N/OFQ system among basal ganglia circuits with particular implications for developmental regulation or wiring of striato-nigral circuits.
Keywords: dendron bouquet; dopamine; nociceptin; opioid; orphanin F/Q; striatum; striosome.
© 2024 The Author(s). The Journal of Comparative Neurology published by Wiley Periodicals LLC.
Figures

Update of
-
Developmental and adult striatal patterning of nociceptin ligand marks striosomal population with direct dopamine projections.bioRxiv [Preprint]. 2024 May 15:2024.05.15.594426. doi: 10.1101/2024.05.15.594426. bioRxiv. 2024. Update in: J Comp Neurol. 2024 Dec;532(12):e70003. doi: 10.1002/cne.70003. PMID: 38798373 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

