Origin and evolution of auxin-mediated acid growth
- PMID: 39656208
- PMCID: PMC11665922
- DOI: 10.1073/pnas.2412493121
Origin and evolution of auxin-mediated acid growth
Abstract
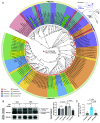
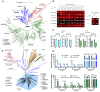
The classical acid growth theory suggests that auxin stimulates cell expansion by triggering apoplast acidification via plasma membrane (PM)-localized H+-ATPase. Here, we reconstructed the origin and evolutionary history of auxin-mediated acid growth. Comparative phylogenomic analysis showed that most core components of acid growth originated in Charophyta and then underwent subclass expansion and functional innovation during plant terrestrialization. In Charophyceae algae Chara braunii, we found that PM H+-ATPase has formed a core regulatory module with TMK and PP2C.D, which can be activated by photosynthesis-dependent phosphorylation through light rather than auxin. Despite the lack of canonical auxin receptor TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX (TIR1/AFB), auxin elicits significant internodal elongation and transcriptional reprogramming in C. braunii, implying the existence of an ancient auxin-mediated growth mechanism. We propose that the evolution of acid growth represents a neofunctional adaptation to terrestrial environments, in which PM H+-ATPase in carbon concentrating for photosynthesis was utilized to acidify apoplast for cell expansion, and the core components responsible for acid growth eventually established a regulatory network in land plants by connecting with the TIR1/AFB pathway.
Keywords: acid growth; auxin; cell expansion; evolution; plant terrestrialization.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- Hager A., Menzel H., Krauss A., Versuche und Hypothese zur Primärwirkung des Auxins beim Streckungswachstum. Planta 100, 47–75 (1971). - PubMed
-
- Claussen M., Lüthe H., Blatt M., Böttger M., Auxin-induced growth and its linkage to potassium channels. Planta 201, 227–234 (1997).
-
- Weijers D., Wagner D., Transcriptional responses to the auxin hormone. Annu. Rev. Plant. Biol. 67, 539–574 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

