Evaluation of VP4-VP2 sequencing for molecular typing of human enteroviruses
- PMID: 39656727
- PMCID: PMC11630573
- DOI: 10.1371/journal.pone.0311806
Evaluation of VP4-VP2 sequencing for molecular typing of human enteroviruses
Abstract
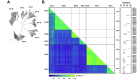
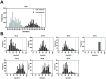
Enteroviruses and rhinoviruses are highly diverse, with over 300 identified types. Reverse transcription-polymerase chain reaction (RT-PCR) assays targeting their VP1, VP4, and partial VP2 (VP4-pVP2) genomic regions are used for detection and identification. The VP4-pVP2 region is particularly sensitive to RT-PCR detection, making it efficient for clinical specimen analysis. However, a standard type identification method using this region is lacking. This study aimed to establish such a method by examining the divergence of VP4-pVP2 amino acid sequences between enterovirus and rhinovirus prototypes. Pairwise analysis of 249 types indicated a 95% threshold for enterovirus intra-species identification but not for rhinovirus prototypes. Protein BLAST search analyses of representative enterovirus prototypes, including EV-A71, EV-D68, CVA6, CVA10, CVA16, and polioviruses (PVs), validated the 95% threshold for typing, with a few exceptions such as PV1-PV2 and CVA6-CVA10, as well as some EV-C types. This study proposes a criterion for typing based on VP4-pVP2 amino acids, which can aid in rapid enterovirus diagnosis during routine clinical or environmental surveillance and emergency outbreaks. Our research confirms the reliability of the suggested VP4-pVP2-based threshold for typing and its potential application in laboratory settings.
Copyright: © 2024 Kitamura, Arita. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Link-Gelles R, Lutterloh E, Schnabel Ruppert P, Backenson PB, St George K, Rosenberg ES, et al.. Public Health Response to a Case of Paralytic Poliomyelitis in an Unvaccinated Person and Detection of Poliovirus in Wastewater—New York, June-August 2022. MMWR Morb Mortal Wkly Rep. 2022;71:1065–1068. doi: 10.15585/mmwr.mm7133e2 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

