The good, the bad, and the hazardous: comparative genomic analysis unveils cell wall features in the pathogen Candidozyma auris typical for both baker's yeast and Candida
- PMID: 39656857
- PMCID: PMC11657238
- DOI: 10.1093/femsyr/foae039
The good, the bad, and the hazardous: comparative genomic analysis unveils cell wall features in the pathogen Candidozyma auris typical for both baker's yeast and Candida
Abstract
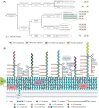
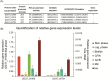
The drug-resistant pathogenic yeast Candidozyma auris (formerly named Candida auris) is considered a critical health problem of global importance. As the cell wall plays a crucial role in pathobiology, here we performed a detailed bioinformatic analysis of its biosynthesis in C. auris and related Candidozyma haemuli complex species using Candida albicans and Saccharomyces cerevisiae as references. Our data indicate that the cell wall architecture described for these reference yeasts is largely conserved in Candidozyma spp.; however, expansions or reductions in gene families point to subtle alterations, particularly with respect to β--1,3--glucan synthesis and remodeling, phosphomannosylation, β-mannosylation, and glycosylphosphatidylinositol (GPI) proteins. In several aspects, C. auris holds a position in between C. albicans and S. cerevisiae, consistent with being classified in a separate genus. Strikingly, among the identified putative GPI proteins in C. auris are adhesins typical for both Candida (Als and Hyr/Iff) and Saccharomyces (Flo11 and Flo5-like flocculins). Further, 26 putative C. auris GPI proteins lack homologs in Candida genus species. Phenotypic analysis of one such gene, QG37_05701, showed mild phenotypes implicating a role associated with cell wall β-1,3-glucan. Altogether, our study uncovered a wealth of information relevant for the pathogenicity of C. auris as well as targets for follow-up studies.
Keywords: Candida auris; GPI proteins; adhesins; candidiasis; cell wall; glucan; mannosylation.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

