Meta learning for mutant HLA class I epitope immunogenicity prediction to accelerate cancer clinical immunotherapy
- PMID: 39656887
- PMCID: PMC11630330
- DOI: 10.1093/bib/bbae625
Meta learning for mutant HLA class I epitope immunogenicity prediction to accelerate cancer clinical immunotherapy
Abstract
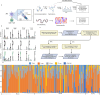
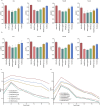
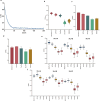
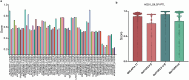
Accurate prediction of binding between human leukocyte antigen (HLA) class I molecules and antigenic peptide segments is a challenging task and a key bottleneck in personalized immunotherapy for cancer. Although existing prediction tools have demonstrated significant results using established datasets, most can only predict the binding affinity of antigenic peptides to HLA and do not enable the immunogenic interpretation of new antigenic epitopes. This limitation results from the training data for the computational models relying heavily on a large amount of peptide-HLA (pHLA) eluting ligand data, in which most of the candidate epitopes lack immunogenicity. Here, we propose an adaptive immunogenicity prediction model, named MHLAPre, which is trained on the large-scale MS-derived HLA I eluted ligandome (mostly presented by epitopes) that are immunogenic. Allele-specific and pan-allelic prediction models are also provided for endogenous peptide presentation. Using a meta-learning strategy, MHLAPre rapidly assessed HLA class I peptide affinities across the whole pHLA pairs and accurately identified tumor-associated endogenous antigens. During the process of adaptive immune response of T-cells, pHLA-specific binding in the antigen presentation is only a pre-task for CD8+ T-cell recognition. The key factor in activating the immune response is the interaction between pHLA complexes and T-cell receptors (TCRs). Therefore, we performed transfer learning on the pHLA model using the pHLA-TCR dataset. In pHLA binding task, MHLAPre demonstrated significant improvement in identifying neoepitope immunogenicity compared with five state-of-the-art models, proving its effectiveness and robustness. After transfer learning of the pHLA-TCR data, MHLAPre also exhibited relatively superior performance in revealing the mechanism of immunotherapy. MHLAPre is a powerful tool to identify neoepitopes that can interact with TCR and induce immune responses. We believe that the proposed method will greatly contribute to clinical immunotherapy, such as anti-tumor immunity, tumor-specific T-cell engineering, and personalized tumor vaccine.
Keywords: HLA genotyping; deep learning; epitope specificity; immunoinformatics; transfer learning.
© The Author(s) 2024. Published by Oxford University Press.
Figures

Similar articles
-
CapHLA: a comprehensive tool to predict peptide presentation and binding to HLA class I and class II.Brief Bioinform. 2024 Nov 22;26(1):bbae595. doi: 10.1093/bib/bbae595. Brief Bioinform. 2024. PMID: 39688477 Free PMC article.
-
Population-level distribution and putative immunogenicity of cancer neoepitopes.BMC Cancer. 2018 Apr 13;18(1):414. doi: 10.1186/s12885-018-4325-6. BMC Cancer. 2018. PMID: 29653567 Free PMC article.
-
ImmuneApp for HLA-I epitope prediction and immunopeptidome analysis.Nat Commun. 2024 Oct 16;15(1):8926. doi: 10.1038/s41467-024-53296-0. Nat Commun. 2024. PMID: 39414796 Free PMC article.
-
Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry.Immunology. 2018 Jul;154(3):331-345. doi: 10.1111/imm.12936. Epub 2018 May 8. Immunology. 2018. PMID: 29658117 Free PMC article. Review.
-
T Cell Epitope Prediction and Its Application to Immunotherapy.Front Immunol. 2021 Sep 15;12:712488. doi: 10.3389/fimmu.2021.712488. eCollection 2021. Front Immunol. 2021. PMID: 34603286 Free PMC article. Review.
Cited by
-
A systematic review of T cell epitopes defined from the proteome of human immunodeficiency virus.Virus Res. 2025 Aug;358:199602. doi: 10.1016/j.virusres.2025.199602. Epub 2025 Jun 23. Virus Res. 2025. PMID: 40562176 Free PMC article. Review.
-
Promising future of breast cancer vaccine asking for multidisciplinary collaboration: a literature review.Front Cell Dev Biol. 2025 Apr 24;13:1578883. doi: 10.3389/fcell.2025.1578883. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40342927 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

