The Sensory Shark: High-quality Morphological, Genomic and Transcriptomic Data for the Small-spotted Catshark Scyliorhinus Canicula Reveal the Molecular Bases of Sensory Organ Evolution in Jawed Vertebrates
- PMID: 39657112
- PMCID: PMC11979771
- DOI: 10.1093/molbev/msae246
The Sensory Shark: High-quality Morphological, Genomic and Transcriptomic Data for the Small-spotted Catshark Scyliorhinus Canicula Reveal the Molecular Bases of Sensory Organ Evolution in Jawed Vertebrates
Abstract
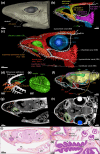
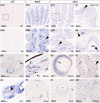
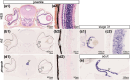
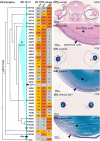
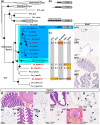
Cartilaginous fishes (chondrichthyans: chimeras and elasmobranchs -sharks, skates, and rays) hold a key phylogenetic position to explore the origin and diversifications of jawed vertebrates. Here, we report and integrate reference genomic, transcriptomic, and morphological data in the small-spotted catshark Scyliorhinus canicula to shed light on the evolution of sensory organs. We first characterize general aspects of the catshark genome, confirming the high conservation of genome organization across cartilaginous fishes, and investigate population genomic signatures. Taking advantage of a dense sampling of transcriptomic data, we also identify gene signatures for all major organs, including chondrichthyan specializations, and evaluate expression diversifications between paralogs within major gene families involved in sensory functions. Finally, we combine these data with 3D synchrotron imaging and in situ gene expression analyses to explore chondrichthyan-specific traits and more general evolutionary trends of sensory systems. This approach brings to light, among others, novel markers of the ampullae of Lorenzini electrosensory cells, a duplication hotspot for crystallin genes conserved in jawed vertebrates, and a new metazoan clade of the transient-receptor potential (TRP) family. These resources and results, obtained in an experimentally tractable chondrichthyan model, open new avenues to integrate multiomics analyses for the study of elasmobranchs and jawed vertebrates.
Keywords: TRP ion channels; cartilagious fish genome; electroreceptors; olfactory receptors; transcriptomics; vertebrate evolution.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures





Update of
-
The sensory shark: high-quality morphological, genomic and transcriptomic data for the small-spotted catshark Scyliorhinus canicula reveal the molecular bases of sensory organ evolution in jawed vertebrates.bioRxiv [Preprint]. 2024 Jul 2:2024.05.23.595469. doi: 10.1101/2024.05.23.595469. bioRxiv. 2024. Update in: Mol Biol Evol. 2024 Dec 6;41(12):msae246. doi: 10.1093/molbev/msae246. PMID: 39005470 Free PMC article. Updated. Preprint.
References
-
- Ballard WW, Mellinger J, Lechenault H. A series of stages for development of Scyliorhinus canicula the lesser spotted dogfish (Chondrichthyes: Scyliorhinidae). J Exp Zool. 1993:267(3):318–336. 10.1002/jez.1402670309. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

