Effects of maternal Escherichia coli lipopolysaccharide exposure on offspring: insights from lncRNA analysis in laying hens
- PMID: 39657467
- PMCID: PMC11681910
- DOI: 10.1016/j.psj.2024.104599
Effects of maternal Escherichia coli lipopolysaccharide exposure on offspring: insights from lncRNA analysis in laying hens
Abstract
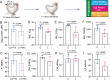
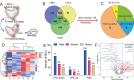
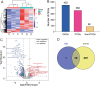
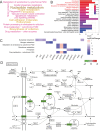
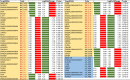
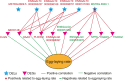
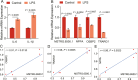
Parental living environment significantly impacts on offspring, yet related studies are lacking in livestock and poultry production. The present study found that lipopolysaccharide (LPS, Escherichia coli, 0.2 mg/kg) stimulation in F0 hens led to growth retardation and a decrease in egg-laying rate in the unchallenged F1 hens. Using strand-specific transcriptomic data of peripheral blood mononuclear cells (PBMCs) in F1 hens, we identified 100 differentially expressed lncRNAs (DELs) and 452 differentially expressed genes (DEGs). LPS primarily affected the metabolic pathways of the offspring, possibly reducing the egg-laying rate of the F1 hens by inhibiting the ferroptosis signaling pathway and the expression of DEGs involved, such as NCOA4, SLC40A1, STEAP3, and TFRC. Using Pearson correlation analysis, we constructed a lncRNA-mRNA-egg-laying rate regulation network and found that the newly identified lncRNA MSTRG.6500.1 and its positively regulated target genes (ENSGALT00000051184, ENSGALT00000053276, NPPA, OSBP2, and TRARG1) were significantly downregulated in the F1 LPS group, which might be the main reason for the decrease in egg-laying rate of the LPS group. These results provide important references for the study of growth and reproductive performance in laying hens, revealing the impact of parental living environment on animal health and production performance, and providing a theoretical basis for future related research and breeding practices.
Keywords: Chicken; Egg-laying rate; LPS maternal stimulation; Offspring; lncRNA.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Andrews, S. 2010. FastQC: a quality control tool for high throughput sequence data. (online).
-
- Ben Maamar M., Sadler-Riggleman I., Beck D., McBirney M., Nilsson E., Klukovich R., Xie Y., Tang C., Yan W., Skinner M.K. Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate vinclozolin-induced epigenetic transgenerational inheritance of disease. Environ. Epigenet. 2018;4:1–19. - PMC - PubMed
-
- Bidne K., Dickson M., Ross J., Baumgard L., Keating A. Disruption of female reproductive function by endotoxins. Reprod. 2018;155:R169–R181. - PubMed
-
- Bradford B., Yuan K., Farney J., Mamedova L., Carpenter A. Invited review: inflammation during the transition to lactation: new adventures with an old flame. J. Dairy Sci. 2015;98:6631–6650. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

