Castrate-resistant prostate cancer response to taxane is determined by an HNF1-dependent apoptosis resistance circuit
- PMID: 39657662
- PMCID: PMC11722106
- DOI: 10.1016/j.xcrm.2024.101868
Castrate-resistant prostate cancer response to taxane is determined by an HNF1-dependent apoptosis resistance circuit
Abstract
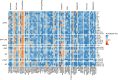
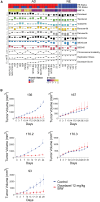
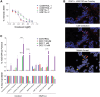
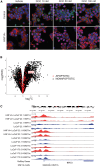
Metastatic castrate-resistant prostate cancer (mCRPC) is a genetically and phenotypically heterogeneous cancer where advancements are needed in biomarker discovery and targeted therapy. A critical and often effective component of treatment includes taxanes. We perform a high-throughput screen across a cohort of 30 diverse patient-derived castrate-resistant prostate cancer (CRPC) organoids to a library of 78 drugs. Combining quantitative response measures with transcriptomic analyses demonstrates that HNF1 homeobox A (HNF1A) drives a transcriptional program of taxane resistance, commonly dependent upon cellular inhibitor of apoptosis protein 2 (cIAP2). Monotherapy with cIAP2 inhibitor LCL161 is sufficient to treat HNF1A+ models of mCRPC previously resistant to docetaxel. These data may be useful in future clinical trial designs.
Keywords: BIRC3; HNF1A; apoptosis; docetaxel; drug resistance; organoid; pharmacology; prostate cancer; xenograft.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests E.C. received sponsored research funding from Sanofi, Gilead, AbbVie, Genentech, Janssen Research, AstraZeneca, GSK, Bayer Pharmaceuticals, Forma Pharmaceuticals, Foghorn, Kronos, and MarcoGenics.
Figures




References
-
- Mohler J.L., Antonarakis E.S., Armstrong A.J., D'Amico A.V., Davis B.J., Dorff T., Eastham J.A., Enke C.A., Farrington T.A., Higano C.S., et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2019;17:479–505. doi: 10.6004/jnccn.2019.0023. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

