Integrated molecular characterization of sarcomatoid hepatocellular carcinoma
- PMID: 39657751
- PMCID: PMC12016616
- DOI: 10.3350/cmh.2024.0686
Integrated molecular characterization of sarcomatoid hepatocellular carcinoma
Abstract
Backgrounds/aims: Sarcomatoid hepatocellular carcinoma (HCC) is a rare histological subtype of HCC characterized by extremely poor prognosis; however, its molecular characterization has not been elucidated.
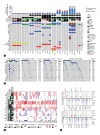
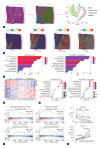
Methods: In this study, we conducted an integrated multiomics study of whole-exome sequencing, RNA-seq, spatial transcriptome, and immunohistochemical analyses of 28 paired sarcomatoid tumor components and conventional HCC components from 10 patients with sarcomatoid HCC, in order to identify frequently altered genes, infer the tumor subclonal architectures, track the genomic evolution, and delineate the transcriptional characteristics of sarcomatoid HCCs.
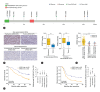
Results: Our results showed that the sarcomatoid HCCs had poor prognosis. The sarcomatoid tumor components and the conventional HCC components were derived from common ancestors, mostly accessing similar mutational processes. Clonal phylogenies demonstrated branched tumor evolution during sarcomatoid HCC development and progression. TP53 mutation commonly occurred at tumor initiation, whereas ARID2 mutation often occurred later. Transcriptome analyses revealed the epithelial-mesenchymal transition (EMT) and hypoxic phenotype in sarcomatoid tumor components, which were confirmed by immunohistochemical staining. Moreover, we identified ARID2 mutations in 70% (7/10) of patients with sarcomatoid HCC but only 1-5% of patients with non-sarcomatoid HCC. Biofunctional investigations revealed that inactivating mutation of ARID2 contributes to HCC growth and metastasis and induces EMT in a hypoxic microenvironment.
Conclusion: We offer a comprehensive description of the molecular basis for sarcomatoid HCC, and identify genomic alteration (ARID2 mutation) together with the tumor microenvironment (hypoxic microenvironment), that may contribute to the formation of the sarcomatoid tumor component through EMT, leading to sarcomatoid HCC development and progression.
Keywords: ARID2; Epithelial–mesenchymal transition; Multiomics; Sarcomatoid hepatocellular carcinoma.
Conflict of interest statement
The authors have no conflicts to disclose.
Figures





References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
-
- Zhou SL, Zhou ZJ, Hu ZQ, Huang XW, Wang Z, Chen EB, et al. Tumor-associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology. 2016;150:1646–1658.e17. - PubMed
-
- Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–1982. - PubMed
-
- Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet 2016;48:500-509. Erratum in: Nat Genet. 2016;48:700. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

