Autoencoder-based phenotyping of ophthalmic images highlights genetic loci influencing retinal morphology and provides informative biomarkers
- PMID: 39657956
- PMCID: PMC11751639
- DOI: 10.1093/bioinformatics/btae732
Autoencoder-based phenotyping of ophthalmic images highlights genetic loci influencing retinal morphology and provides informative biomarkers
Abstract
Motivation: Genome-wide association studies (GWAS) have been remarkably successful in identifying associations between genetic variants and imaging-derived phenotypes. To date, the main focus of these analyses has been on established, clinically-used imaging features. We sought to investigate if deep learning approaches can detect more nuanced patterns of image variability.
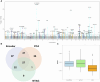
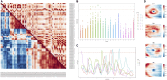
Results: We used an autoencoder to represent retinal optical coherence tomography (OCT) images from 31 135 UK Biobank participants. For each subject, we obtained a 64-dimensional vector representing features of retinal structure. GWAS of these autoencoder-derived imaging parameters identified 118 statistically significant loci; 41 of these associations were also significant in a replication study. These loci encompassed variants previously linked with retinal thickness measurements, ophthalmic disorders, and/or neurodegenerative conditions. Notably, the generated retinal phenotypes were found to contribute to predictive models for glaucoma and cardiovascular disorders. Overall, we demonstrate that self-supervised phenotyping of OCT images enhances the discoverability of genetic factors influencing retinal morphology and provides epidemiologically informative biomarkers.
Availability and implementation: Code and data links available at https://github.com/tf2/autoencoder-oct.
© The Author(s) 2024. Published by Oxford University Press.
Figures



References
-
- Budu-Aggrey A, Hysi P, Kehoe PG et al. The relationship between open angle glaucoma, optic disc morphology and Alzheimer’s disease: a Mendelian randomization study. bioRxiv 2020; 10.1101/2020.08.30.20184846, preprint: not peer reviewed. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

