Long G4-rich enhancers target promoters via a G4 DNA-based mechanism
- PMID: 39658038
- PMCID: PMC11754661
- DOI: 10.1093/nar/gkae1180
Long G4-rich enhancers target promoters via a G4 DNA-based mechanism
Abstract
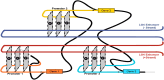
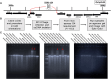
Several studies have now described instances where G-rich sequences in promoters and enhancers regulate gene expression through forming G-quadruplex (G4) structures. Relatedly, our group recently identified 301 long genomic stretches significantly enriched for minimal G4 motifs (LG4s) in humans and found the majority of these overlap annotated enhancers, and furthermore, that the promoters regulated by these LG4 enhancers are similarly enriched with G4-capable sequences. While the generally accepted model for enhancer:promoter specificity maintains that interactions are dictated by enhancer- and promoter-bound transcriptional activator proteins, the current study tested an alternative hypothesis: that LG4 enhancers interact with cognate promoters via a direct G4:G4 DNA-based mechanism. This work establishes the nuclear proximity of LG4 enhancer:promoter pairs, biochemically demonstrates the ability of individual LG4 single-stranded DNAs (ssDNAs) to directly interact target promoter ssDNAs, and confirms that these interactions, as well as the ability of LG4 enhancers to activate target promoters in culture, are mediated by G4 DNA.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

