Integration of lipidomics with targeted, single cell, and spatial transcriptomics defines an unresolved pro-inflammatory state in colon cancer
- PMID: 39658263
- PMCID: PMC11885055
- DOI: 10.1136/gutjnl-2024-332535
Integration of lipidomics with targeted, single cell, and spatial transcriptomics defines an unresolved pro-inflammatory state in colon cancer
Abstract
Background: Over a century ago, Virchow proposed that cancer represents a chronically inflamed, poorly healing wound. Normal wound healing is represented by a transitory phase of inflammation, followed by a pro-resolution phase, with prostaglandin (PGE2/PGD2)-induced 'lipid class switching' producing inflammation-quenching lipoxins (LXA4, LXB4).
Objective: We explored if lipid dysregulation in colorectal cancers (CRCs) is driven by a failure to resolve inflammation.
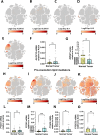
Design: We performed liquid chromatography and tandem mass spectrometry (LC-MS/MS) untargeted analysis of 40 human CRC and normal paired samples and targeted, quantitative analysis of 81 human CRC and normal paired samples. We integrated analysis of lipidomics, quantitative reverse transcription-PCR, large scale gene expression, and spatial transcriptomics with public scRNASEQ data to characterize pattern, expression and cellular localisation of genes that produce and modify lipid mediators.
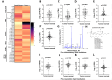
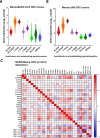
Results: Targeted, quantitative LC-MS/MS demonstrated a marked imbalance of pro-inflammatory mediators, with a dearth of resolving lipid mediators. In tumours, we observed prominent over-expression of arachidonic acid derivatives, the genes encoding their synthetic enzymes and receptors, but poor expression of genes producing pro-resolving synthetic enzymes and resultant lipoxins (LXA4, LXB4) and associated receptors. These results indicate that CRC is the product of defective lipid class switching likely related to inadequate or ineffective levels of PGE2/PGD2.
Conclusion: We show that the lipidomic profile of CRC tumours exhibits a distinct pro-inflammatory bias with a deficiency of endogenous resolving mediators secondary to defective lipid class switching. These observations pave the way for 'resolution medicine', a novel therapeutic approach for inducing or providing resolvins to mitigate the chronic inflammation driving cancer growth and progression.
Keywords: COLORECTAL CANCER; EICOSANOIDS; GENE EXPRESSION; INFLAMMATION; LIPID METABOLISM.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
