Protein import into bacterial endosymbionts and evolving organelles
- PMID: 39658314
- PMCID: PMC12176269
- DOI: 10.1111/febs.17356
Protein import into bacterial endosymbionts and evolving organelles
Abstract
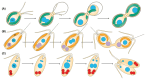
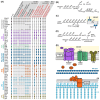
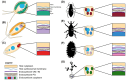
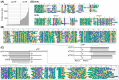
Bacterial endosymbionts are common throughout the eukaryotic tree of life and provide a range of essential functions. The intricate integration of bacterial endosymbionts into a host led to the formation of the energy-converting organelles, mitochondria and plastids, that have shaped eukaryotic evolution. Protein import from the host has been regarded as one of the distinguishing features of organelles as compared to endosymbionts. In recent years, research has delved deeper into a diverse range of endosymbioses and discovered evidence for 'exceptional' instances of protein import outside of the canonical organelles. Here we review the current evidence for protein import into bacterial endosymbionts. We cover both 'recently evolved' organelles, where there is evidence for hundreds of imported proteins, and endosymbiotic systems where currently only single protein import candidates are described. We discuss the challenges of establishing protein import machineries and the diversity of mechanisms that have independently evolved to solve them. Understanding these systems and the different independent mechanisms, they have evolved is critical to elucidate how cellular integration arises and deepens at the endosymbiont to organelle interface. We finish by suggesting approaches that could be used in the future to address the open questions. Overall, we believe that the evidence now suggests that protein import into bacterial endosymbionts is more common than generally realized, and thus that there is an increasing number of partnerships that blur the distinction between endosymbiont and organelle.
Keywords: Paulinella; Strigomonadinae; UCYN‐A; endosymbiosis; envelope membranes; mealybug; nitroplast; organellogenesis; protein translocation; targeting signals.
© 2024 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M & Penny D (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99, 12246–12251. - PMC - PubMed
-
- Timmis JN, Ayliffe MA, Huang CY & Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5, 123–135. - PubMed
-
- Nowack ECM & Weber APM (2018) Genomics‐informed insights into endosymbiotic organelle evolution in photosynthetic eukaryotes. Annu Rev Plant Biol 69, 51–84. - PubMed
-
- Roger AJ, Muñoz‐Gómez SA & Kamikawa R (2017) The origin and diversification of mitochondria. Curr Biol 27, R1177–R1192. - PubMed
-
- Chen LJ & Li HM (2017) Stable megadalton TOC–TIC supercomplexes as major mediators of protein import into chloroplasts. Plant J 92, 178–188. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

