Heterogeneity-driven phenotypic plasticity and treatment response in branched-organoid models of pancreatic ductal adenocarcinoma
- PMID: 39658630
- PMCID: PMC12176653
- DOI: 10.1038/s41551-024-01273-9
Heterogeneity-driven phenotypic plasticity and treatment response in branched-organoid models of pancreatic ductal adenocarcinoma
Abstract
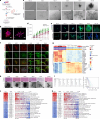
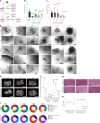
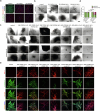
In patients with pancreatic ductal adenocarcinoma (PDAC), intratumoural and intertumoural heterogeneity increases chemoresistance and mortality rates. However, such morphological and phenotypic diversities are not typically captured by organoid models of PDAC. Here we show that branched organoids embedded in collagen gels can recapitulate the phenotypic landscape seen in murine and human PDAC, that the pronounced molecular and morphological intratumoural and intertumoural heterogeneity of organoids is governed by defined transcriptional programmes (notably, epithelial-to-mesenchymal plasticity), and that different organoid phenotypes represent distinct tumour-cell states with unique biological features in vivo. We also show that phenotype-specific therapeutic vulnerabilities and modes of treatment-induced phenotype reprogramming can be captured in phenotypic heterogeneity maps. Our methodology and analyses of tumour-cell heterogeneity in PDAC may guide the development of phenotype-targeted treatment strategies.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: M.L. was a contractor for the Chan Zuckerberg Initiative and received speaker fees from Pfizer and Janssen Pharmaceuticals. F.J.T. consults for Immunai Inc., Singularity Bio B.V., CytoReason Ltd, Cellarity and Omniscope, and has ownership interest in Dermagnostix GmbH and Cellarity. The other authors declare no competing interests.
Figures














References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

