Efficient genome monomer higher-order structure annotation and identification using the GRMhor algorithm
- PMID: 39659587
- PMCID: PMC11630843
- DOI: 10.1093/bioadv/vbae191
Efficient genome monomer higher-order structure annotation and identification using the GRMhor algorithm
Abstract
Motivation: Tandem monomeric units, integral components of eukaryotic genomes, form higher-order repeat (HOR) structures that play crucial roles in maintaining chromosome integrity and regulating gene expression and protein abundance. Given their significant influence on processes such as evolution, chromosome segregation, and disease, developing a sensitive and automated tool for identifying HORs across diverse genomic sequences is essential.
Results: In this study, we applied the GRMhor (Global Repeat Map hor) algorithm to analyse the centromeric region of chromosome 20 in three individual human genomes, as well as in the centromeric regions of three higher primates. In all three human genomes, we identified six distinct HOR arrays, which revealed significantly greater differences in the number of canonical and variant copies, as well as in their overall structure, than would be expected given the 99.9% genetic similarity among humans. Furthermore, our analysis of higher primate genomes, which revealed entirely different HOR sequences, indicates a much larger genomic divergence between humans and higher primates than previously recognized. These results underscore the suitability of the GRMhor algorithm for studying specificities in individual genomes, particularly those involving repetitive monomers in centromere structure, which is essential for proper chromosome segregation during cell division, while also highlighting its utility in exploring centromere evolution and other repetitive genomic regions.
Availability and implementation: Source code and example binaries freely available for download at github.com/gluncic/GRM2023.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
All authors of this article declare that they have no conflicts of interest.
Figures
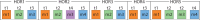
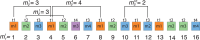





References
-
- Alexandrov I, Kazakov A, Tumeneva I. et al. Alpha-satellite DNA of primates: old and new families. Chromosoma 2001;110:253–66. - PubMed
-
- Alexandrov IA, Mashkova TD, Akopian TA. et al. Chromosome-specific alpha satellites: two distinct families on human chromosome 18. Genomics 1991;11:15–23. - PubMed
-
- Altschul SF, Gish W, Miller W. et al. Basic local alignment search tool. J Mol Biol 1990;215:403–10. - PubMed
LinkOut - more resources
Full Text Sources

