Genomic tumor evolution dictates human medulloblastoma progression
- PMID: 39659836
- PMCID: PMC11629688
- DOI: 10.1093/noajnl/vdae172
Genomic tumor evolution dictates human medulloblastoma progression
Abstract
Background: Medulloblastoma (MB) is the most common high-grade pediatric brain tumor, comprised of 4 main molecular subgroups-sonic-hedgehog (SHH), Wnt, Group 3, and Group 4. Group 3 and Group 4 tumors are the least characterized MB subgroups, despite Group 3 having the worst prognosis (~50% survival rate), and Group 4 being the most prevalent. Such poor characterization can be attributed to high levels of inter- and intratumoral heterogeneity, making it difficult to identify common therapeutic targets.
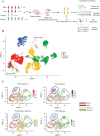
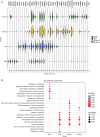
Methods: In this study, we generated single-cell sequencing data from 14 MB patients spanning all subgroups that we complemented with publicly available single-cell data from Group 3 patients. We used a ligand-receptor analysis tool (CellChat), expression- and allele-based copy-number variation (CNV) detection methods, and RNA velocity analysis to characterize tumor cell-cell interactions, established a connection between CNVs and temporal tumor progression, and unraveled tumor evolution.
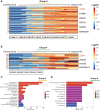
Results: We show that MB tumor cells follow a temporal trajectory from those with low CNV levels to those with high CNV levels, allowing us to identify early and late markers for SHH, Group 3, and Group 4 MBs. Our study also identifies SOX4 upregulation as a major event in later tumor clones for Group 3 and Group 4 MBs, suggesting it as a potential therapeutic target for both subgroups.
Conclusion: Taken together, our findings highlight MB's inherent tumor heterogeneity and offer promising insights into potential drivers of MB tumor evolution particularly in Group 3 and Group 4 MBs.
Keywords: medulloblastoma; single-cell sequencing; tumor heterogeneity.
© The Author(s) 2024. Published by Oxford University Press, the Society for Neuro-Oncology and the European Association of Neuro-Oncology.
Conflict of interest statement
All authors declare no conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous
