Bioinformatics- and quantitative proteomics-based identification of gastric adenocarcinoma-related proteins and analysis
- PMID: 39659938
- PMCID: PMC11626274
- DOI: 10.62347/BVFO4627
Bioinformatics- and quantitative proteomics-based identification of gastric adenocarcinoma-related proteins and analysis
Abstract
Background: The emergence of immune resistance and a lack of effective therapeutic targets have become significant challenges in immunotherapy, highlighting the urgent need for new molecular markers and treatment targets. Moreover, the significance and mechanisms of PGRN (Progranulin) in gastric cancer remain ambiguous.
Objective: To identify differentially expressed proteins in gastric cancer and elucidate the function and mechanism of PGRN.
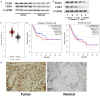
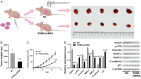
Methods: The data-independent acquisition proteomics was used to identify the differentially expressed proteins in gastric adenocarcinoma and the corresponding paraneoplastic tissues, providing a comprehensive dataset of gastric cancer-related proteins. The function and mechanism of PGRN in gastric cancer were further explored using a series of experiments, including RT-qPCR (Real Time-Quantitative Polymerase Chain Reaction), cell transfection, cell viability assays, cell scratch, immunohistochemistry and Transwell assays, Western blot, and a mouse tumor-bearing model. These investigations were combined with bioinformatics analyses to examine the relationship between PGRN expression and clinical-pathological characteristics, confirming its high expression of PGRN in gastric cancer tissues.
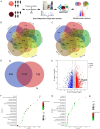
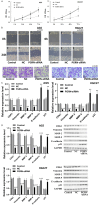
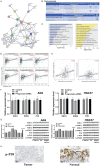
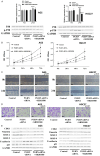
Results: We identified a large number of differentially expressed proteins between gastric cancer and adjacent tissues and conducted an initial functional analysis. Further studies on PGRN showed that it was associated with gastric cancer prognosis and lymph node metastasis. The inhibition of PGRN expression led to reduced cell viability, migration, and invasion, with corresponding changes in related genes and proteins. In a mouse tumor-bearing model, the tumor growth of the subcutaneously transplanted tumors in nude mice was reduced after the inhibition of PGRN expression. An in-depth functional analysis of PGRN was performed using bioinformatics to predict protein interactions, miRNA regulation, and relationships with multiple immune cell types. Enrichment analysis indicated that PGRN is involved in multiple signaling pathways, with the MAPK (Mitogen-Activated Protein Kinase) pathway selected for validation. In AGS and HGC27 cells, PGRN inhibition led to increased expression of phosphorylated p38 (p-p38) in the MAPK pathway, suggesting that PGRN may promote gastric cancer development by regulating p-p38.
Conclusions: This study identified significant differences in protein expression between gastric adenocarcinoma and adjacent tissues, with PGRN emerging as a key protein influencing gastric cancer proliferation, migration, and invasion. These findings suggest that PGRN could serve as a potential therapeutic target for gastric cancer.
Keywords: Gastric cancer; PGRN; bioinformatics; functional analysis; quantitative proteomics.
AJCR Copyright © 2024.
Conflict of interest statement
None.
Figures
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Dong L, Lu D, Chen R, Lin Y, Zhu H, Zhang Z, Cai S, Cui P, Song G, Rao D, Yi X, Wu Y, Song N, Liu F, Zou Y, Zhang S, Zhang X, Wang X, Qiu S, Zhou J, Wang S, Zhang X, Shi Y, Figeys D, Ding L, Wang P, Zhang B, Rodriguez H, Gao Q, Gao D, Zhou H, Fan J. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell. 2022;40:70–87. e15. - PubMed
LinkOut - more resources
Full Text Sources
