The genome sequence of a woodlouse fly, Melanophora roralis (Linnaeus, 1758)
- PMID: 39659996
- PMCID: PMC11628945
- DOI: 10.12688/wellcomeopenres.23295.1
The genome sequence of a woodlouse fly, Melanophora roralis (Linnaeus, 1758)
Abstract
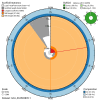
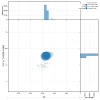
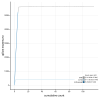
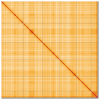
We present a genome assembly from an individual female woodlouse fly, Melanophora roralis (Arthropoda; Insecta; Diptera; Rhinophoridae). The genome sequence has a total length of 565.10 megabases. Most of the assembly (98.9%) is scaffolded into 6 chromosomal pseudomolecules, including the X sex chromosome. The mitochondrial genome has also been assembled and is 19.44 kilobases in length. Gene annotation of this assembly on Ensembl identified 20,321 protein-coding genes.
Keywords: Diptera; Melanophora roralis; chromosomal; genome sequence; woodlouse fly.
Copyright: © 2024 Mitchell R et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

References
-
- Bates A, Clayton-Lucey I, Howard C: Sanger Tree of Life HMW DNA fragmentation: diagenode Megaruptor ®3 for LI PacBio. protocols.io. 2023. 10.17504/protocols.io.81wgbxzq3lpk/v1 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

