Exploring sperm cell motion dynamics: Insights from genetic algorithm-based analysis
- PMID: 39660215
- PMCID: PMC11630665
- DOI: 10.1016/j.csbj.2024.06.008
Exploring sperm cell motion dynamics: Insights from genetic algorithm-based analysis
Abstract
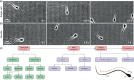
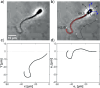
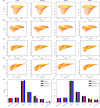
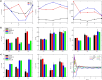
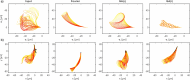
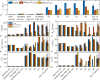
Accurate analysis of sperm cell flagellar dynamics plays a crucial role in understanding sperm motility as flagella parameters determine cell behavior in the spatiotemporal domain. In this study, we introduce a novel approach by harnessing Genetic Algorithms (GA) to analyze sperm flagellar motion characteristics and compare the results with the traditional decomposition method based on Fourier analysis. Our analysis focuses on extracting key parameters of the equation approximating flagellar shape, including beating period time, bending amplitude, mean curvature, and wavelength. Additionally, we delve into the extraction of phase constants and initial swimming directions, vital for the comprehensive study of sperm cell pairs and bundling phenomena. One significant advantage of GA over Fourier analysis is its ability to integrate sperm cell motion data, enabling a more comprehensive analysis. In contrast, Fourier analysis neglects sperm cell motion by transitioning to a sperm-centered coordinate system (material system). In our comparative study, GA consistently outperform the Fourier analysis-based method, yielding a remarkable reduction in fitting error of up to 70% and on average by 45%. An in-depth exploration of the sperm cell motion becomes indispensable in a wide range of applications from complexities of reproductive biology and medicine, to developing soft flagellated microrobots.
Keywords: Biological motion; Flagellum deformation; Genetic algorithm; Motion analysis; Sperm cell dynamics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

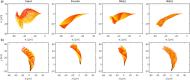
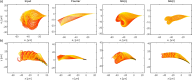
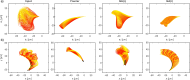
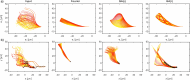
Similar articles
-
Unraveling the Kinematics of Sperm Motion by Reconstructing the Flagellar Wave Motion in 3D.Small Methods. 2022 Mar;6(3):e2101089. doi: 10.1002/smtd.202101089. Epub 2022 Feb 9. Small Methods. 2022. PMID: 35138044
-
Emergent three-dimensional sperm motility: coupling calcium dynamics and preferred curvature in a Kirchhoff rod model.Math Med Biol. 2019 Dec 4;36(4):439-469. doi: 10.1093/imammb/dqy015. Math Med Biol. 2019. PMID: 30325451
-
Microfluidic sperm trap array for single-cell flagellar analysis with unrestricted 2D flagellar movement.Lab Chip. 2024 Oct 9;24(20):4827-4842. doi: 10.1039/d4lc00515e. Lab Chip. 2024. PMID: 39291409
-
Digital image analysis of flagellar beating and microtubule sliding of activated and hyperactivated sperm flagella.Soc Reprod Fertil Suppl. 2007;65:327-30. Soc Reprod Fertil Suppl. 2007. PMID: 17644972 Review.
-
Microscopic analysis of sperm movement: links to mechanisms and protein components.Microscopy (Oxf). 2018 Jun 1;67(3):144-155. doi: 10.1093/jmicro/dfy021. Microscopy (Oxf). 2018. PMID: 29741637 Review.
Cited by
-
Deep Learning Models for Multi-Part Morphological Segmentation and Evaluation of Live Unstained Human Sperm.Sensors (Basel). 2025 May 14;25(10):3093. doi: 10.3390/s25103093. Sensors (Basel). 2025. PMID: 40431886 Free PMC article.
References
-
- Dai C., Zhang Z., Huang J., Wang X., Ru C., Pu H., et al. Automated non-invasive measurement of single sperm's motility and morphology. IEEE Trans Med Imaging. 2018;37(10):2257–2265. - PubMed
LinkOut - more resources
Full Text Sources
