Translation of circular RNAs
- PMID: 39660652
- PMCID: PMC11724312
- DOI: 10.1093/nar/gkae1167
Translation of circular RNAs
Abstract
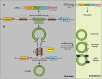
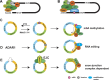
Circular RNAs (circRNAs) are covalently closed RNAs that are present in all eukaryotes tested. Recent RNA sequencing (RNA-seq) analyses indicate that although generally less abundant than messenger RNAs (mRNAs), over 1.8 million circRNA isoforms exist in humans, much more than the number of currently known mRNA isoforms. Most circRNAs are generated through backsplicing that depends on pre-mRNA structures, which are influenced by intronic elements, for example, primate-specific Alu elements, leading to species-specific circRNAs. CircRNAs are mostly cytosolic, stable and some were shown to influence cells by sequestering miRNAs and RNA-binding proteins. We review the increasing evidence that circRNAs are translated into proteins using several cap-independent translational mechanisms, that include internal ribosomal entry sites, N6-methyladenosine RNA modification, adenosine to inosine RNA editing and interaction with the eIF4A3 component of the exon junction complex. CircRNAs are translated under conditions that favor cap-independent translation, notably in cancer and generate proteins that are shorter than mRNA-encoded proteins, which can acquire new functions relevant in diseases.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B.. Scrambled exons. Cell. 1991; 64:607–613. - PubMed
-
- Cocquerelle C., Mascrez B., Hetuin D., Bailleul B.. Mis-splicing yields circular RNA molecules. FASEB J. 1993; 7:155–160. - PubMed
-
- Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R.. Circular transcripts of the testis-determining gene sry in adult mouse testis. Cell. 1993; 73:1019–1030. - PubMed
-
- Kjems J., Garrett R.A.. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcusmobilis. Cell. 1988; 54:693–703. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

