Recent Progress in Modeling and Simulation of Biomolecular Crowding and Condensation Inside Cells
- PMID: 39660892
- PMCID: PMC11683874
- DOI: 10.1021/acs.jcim.4c01520
Recent Progress in Modeling and Simulation of Biomolecular Crowding and Condensation Inside Cells
Abstract
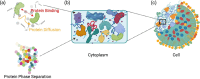
Macromolecular crowding in the cellular cytoplasm can potentially impact diffusion rates of proteins, their intrinsic structural stability, binding of proteins to their corresponding partners as well as biomolecular organization and phase separation. While such intracellular crowding can have a large impact on biomolecular structure and function, the molecular mechanisms and driving forces that determine the effect of crowding on dynamics and conformations of macromolecules are so far not well understood. At a molecular level, computational methods can provide a unique lens to investigate the effect of macromolecular crowding on biomolecular behavior, providing us with a resolution that is challenging to reach with experimental techniques alone. In this review, we focus on the various physics-based and data-driven computational methods developed in the past few years to investigate macromolecular crowding and intracellular protein condensation. We review recent progress in modeling and simulation of biomolecular systems of varying sizes, ranging from single protein molecules to the entire cellular cytoplasm. We further discuss the effects of macromolecular crowding on different phenomena, such as diffusion, protein-ligand binding, and mechanical and viscoelastic properties, such as surface tension of condensates. Finally, we discuss some of the outstanding challenges that we anticipate the community addressing in the next few years in order to investigate biological phenomena in model cellular environments by reproducing in vivo conditions as accurately as possible.
Keywords: Brownian dynamics simulations; cellular cytoplasm; macromolecular crowding; molecular dynamics simulations; phase separation; protein condensation.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Macromolecular Crowding, Phase Separation, and Homeostasis in the Orchestration of Bacterial Cellular Functions.Chem Rev. 2024 Feb 28;124(4):1899-1949. doi: 10.1021/acs.chemrev.3c00622. Epub 2024 Feb 8. Chem Rev. 2024. PMID: 38331392 Free PMC article. Review.
-
Liquid-Liquid Phase Separation in the Presence of Macromolecular Crowding and State-dependent Kinetics.Int J Mol Sci. 2021 Jun 22;22(13):6675. doi: 10.3390/ijms22136675. Int J Mol Sci. 2021. PMID: 34206440 Free PMC article.
-
Thermodynamics of Macromolecular Association in Heterogeneous Crowding Environments: Theoretical and Simulation Studies with a Simplified Model.J Phys Chem B. 2016 Nov 23;120(46):11856-11865. doi: 10.1021/acs.jpcb.6b06243. Epub 2016 Nov 15. J Phys Chem B. 2016. PMID: 27797534 Free PMC article.
-
Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm.Elife. 2016 Nov 1;5:e19274. doi: 10.7554/eLife.19274. Elife. 2016. PMID: 27801646 Free PMC article.
-
Macromolecular crowding in biological systems: hydrodynamics and NMR methods.J Mol Recognit. 2004 Sep-Oct;17(5):397-407. doi: 10.1002/jmr.694. J Mol Recognit. 2004. PMID: 15362098 Review.
Cited by
-
Underestimated role of macromolecular crowding in bioengineered in vitro models of health and diseases.Mater Today Bio. 2025 Apr 17;32:101772. doi: 10.1016/j.mtbio.2025.101772. eCollection 2025 Jun. Mater Today Bio. 2025. PMID: 40331149 Free PMC article.
-
A coarse-grained model for disordered proteins under crowded conditions.Protein Sci. 2025 Aug;34(8):e70232. doi: 10.1002/pro.70232. Protein Sci. 2025. PMID: 40671581 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

