Exploring aggregation genes in a P. aeruginosa chronic infection model
- PMID: 39660900
- PMCID: PMC11784459
- DOI: 10.1128/jb.00429-24
Exploring aggregation genes in a P. aeruginosa chronic infection model
Abstract
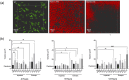
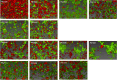
Bacterial aggregates are observed in both natural and artificial environments. In the context of disease, aggregates have been isolated from chronic and acute infections. Pseudomonas aeruginosa (Pa) aggregates contribute significantly to chronic infections, particularly in the lungs of people with cystic fibrosis (CF). Unlike the large biofilm structures observed in vitro, Pa in CF sputum forms smaller aggregates (~10-1,000 cells), and the mechanisms behind their formation remain underexplored. This study aims to identify genes essential and unique to Pa aggregate formation in a synthetic CF sputum media (SCFM2). We cultured Pa strain PAO1 in SCFM2 and LB, both with and without mucin, and used RNA sequencing (RNA-seq) to identify differentially expressed genes. The presence of mucin revealed 13 significantly differentially expressed (DE) genes, predominantly downregulated, with 40% encoding hypothetical proteins unique to aggregates. Using high-resolution microscopy, we assessed the ability of mutants to form aggregates. Notably, no mutant exhibited a completely planktonic phenotype. Instead, we identified multiple spatial phenotypes described as "normal," "entropic," or "impaired." Entropic mutants displayed tightly packed, raft-like structures, while impaired mutants had loosely packed cells. Predictive modeling linked the prioritized genes to metabolic shifts, iron acquisition, surface modification, and quorum sensing. Co-culture experiments with wild-type PAO1 revealed further spatial heterogeneity and the ability to "rescue" some mutant phenotypes, suggesting cooperative interactions during growth. This study enhances our understanding of Pa aggregate biology, specifically the genes and pathways unique to aggregation in CF-like environments. Importantly, it provides insights for developing therapeutic strategies targeting aggregate-specific pathways.
Importance: This study identifies genes essential for the formation of Pseudomonas aeruginosa (Pa) aggregates in cystic fibrosis (CF) sputum, filling a critical gap in understanding their specific biology. Using a synthetic CF sputum model (SCFM2) and RNA sequencing, 13 key genes were identified, whose disruption led to distinct spatial phenotypes observed through high-resolution microscopy. The addition of wild-type cells either rescued the mutant phenotype or increased spatial heterogeneity, suggesting cooperative interactions are involved in aggregate formation. This research advances our knowledge of Pa aggregate biology, particularly the unique genes and pathways involved in CF-like environments, offering valuable insights for developing targeted therapeutic strategies against aggregate-specific pathways.
Keywords: CF; Pseudomonas aeruginosa; aggregate; chronic infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


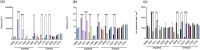
Similar articles
-
Tools for the Real-Time Assessment of a Pseudomonas aeruginosa Infection Model.J Vis Exp. 2021 Apr 6;(170). doi: 10.3791/62420. J Vis Exp. 2021. PMID: 33900293
-
Effect of Shear Stress on Pseudomonas aeruginosa Isolated from the Cystic Fibrosis Lung.mBio. 2016 Aug 2;7(4):e00813-16. doi: 10.1128/mBio.00813-16. mBio. 2016. PMID: 27486191 Free PMC article.
-
Phage Inhibit Pathogen Dissemination by Targeting Bacterial Migrants in a Chronic Infection Model.mBio. 2017 Apr 4;8(2):e00240-17. doi: 10.1128/mBio.00240-17. mBio. 2017. PMID: 28377527 Free PMC article.
-
Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung.Curr Top Microbiol Immunol. 2013;358:91-118. doi: 10.1007/82_2011_199. Curr Top Microbiol Immunol. 2013. PMID: 22311171 Review.
-
Microenvironmental characteristics and physiology of biofilms in chronic infections of CF patients are strongly affected by the host immune response.APMIS. 2017 Apr;125(4):276-288. doi: 10.1111/apm.12668. APMIS. 2017. PMID: 28407427 Review.
References
-
- Kolpen M, Kragh KN, Enciso JB, Faurholt-Jepsen D, Lindegaard B, Egelund GB, Jensen AV, Ravn P, Mathiesen IHM, Gheorge AG, Hertz FB, Qvist T, Whiteley M, Jensen PØ, Bjarnsholt T. 2022. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax 77:1015–1022. doi:10.1136/thoraxjnl-2021-217576 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

