Bacteria-specific modified nucleoside is released and elevated in urine of patients with bacterial infections
- PMID: 39660929
- PMCID: PMC11708014
- DOI: 10.1128/mbio.03124-24
Bacteria-specific modified nucleoside is released and elevated in urine of patients with bacterial infections
Abstract
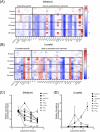
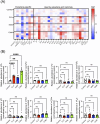
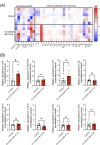
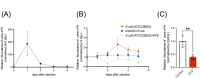
Over 170 types of chemical modifications have been identified in cellular RNAs across the three domains of life. Modified RNA is eventually degraded to constituent nucleosides, and in mammals, modified nucleosides are released into the extracellular space. By contrast, the fate of modified nucleosides in bacteria remains unknown. In this study, we performed liquid chromatography-mass spectroscopy (LC-MS) analysis of modified nucleosides from the RNA of 23 pathogenic bacteria, revealing 2-methyladenosine (m2A) as a common bacteria-specific modified nucleoside detected in all bacterial RNAs. Under normal culture conditions, bacteria did not actively release most modified nucleoside species, but robustly released nucleosides, including m2A, following addition of antibiotics or immune cells. These results indicate that m2A is released following bacterial lysis. Intraperitoneal injection of mice with m2A increased detectable levels of m2A in the urine, indicating that mammals can effectively excrete m2A. Additionally, mice infected with wild-type E. coli showed higher levels of m2A in their urine than mice infected by m2A-deficient rlmN KO E. coli. This suggests that m2A from the infected bacteria is excreted in the urine. Lastly, clinical studies using urine samples from febrile patients revealed significantly elevated levels of m2A during bacterial infections, and these values did not correlate with inflammation severity markers, such as white blood count (WBC) and C-reactive protein (CRP). This study reports the mammalian metabolism of modified nucleosides derived from bacterial RNA, and the elevation of urinary m2A in patients with bacterial infections.
Importance: This study reveals the differences in the fate and release of modified nucleosides in bacteria and mammals. Additionally, our study highlights that external bacteria-damaging factors, such as antibiotics and phagocytosis by host immune cells, promote the release of bacteria-specific modified nucleosides. Furthermore, we found that m2A was elevated in the urine from animal models of bacterial infection and the urine of patients with bacterial infections. Collectively, this work spans basic biology and clinical science, offering valuable insights into the fate of modified nucleosides in bacterial systems and their relevance to infectious diseases.
Keywords: LC-MS; RNA modification; bacterial infection; biomarker; modified nucleoside.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, Yamagata K, Suzuki T, Tomizawa K. 2011. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest 121:3598–3608. doi: 10.1172/JCI58056 - DOI - PMC - PubMed
-
- Wei FY, Zhou B, Suzuki T, Miyata K, Ujihara Y, Horiguchi H, Takahashi N, Xie P, Michiue H, Fujimura A, Kaitsuka T, Matsui H, Koga Y, Mohri S, Suzuki T, Oike Y, Tomizawa K. 2015. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab 21:428–442. doi: 10.1016/j.cmet.2015.01.019 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
