Single-cell epigenetic and clonal analysis decodes disease progression in pediatric acute myeloid leukemia
- PMID: 39661948
- PMCID: PMC11923433
- DOI: 10.1182/blood.2024025618
Single-cell epigenetic and clonal analysis decodes disease progression in pediatric acute myeloid leukemia
Abstract
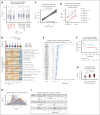
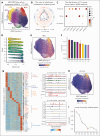
Pediatric acute myeloid leukemia (pAML) is a clonal disease with recurrent genetic alterations that affect epigenetic states. However, the implications of epigenetic dysregulation in disease progression remain unclear. Here, we interrogated single-cell and clonal level chromatin accessibility of bone marrow samples from 28 patients with pAML representing multiple subtypes using mitochondrial single-cell assay for transposase-accessible chromatin with sequencing, which revealed distinct differentiation hierarchies and abnormal chromatin accessibility in a subtype-specific manner. Innate immune signaling was commonly enhanced across subtypes and related to improved advantage of clonal competition and unfavorable prognosis, with further reinforcement in a relapse-associated leukemia stem cell-like population. We identified a panel of 31 innate immunity-related genes to improve the risk classification of patients with pAML. By comparing paired diagnosis and postchemotherapy relapse samples, we showed that primitive cells significantly reduced major histocompatibility complex class II signaling, suggesting an immune evasion mechanism to facilitate their expansion at relapse. Key regulators orchestrating cell cycle dysregulation were identified to contribute to pAML relapse in drug-resistant clones. Our work establishes the single-cell chromatin accessibility landscape at clonal resolution and reveals the critical involvement of epigenetic disruption, offering insights into classification and targeted therapies of patients with pAML.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Review statement: Berthold Göttgens is an Associate Editor of
The current affiliation for B.C. is Department of Medical Technology, Anhui Medical College, Hefei, Anhui, China.
Figures




Comment in
-
The chromatin accessibility landscape of pediatric AML.Blood. 2025 Mar 13;145(11):1109-1111. doi: 10.1182/blood.2024027585. Blood. 2025. PMID: 40080008 No abstract available.
References
-
- Wiggers CRM, Baak ML, Sonneveld E, Nieuwenhuis EES, Bartels M, Creyghton MP. AML subtype is a major determinant of the association between prognostic gene expression signatures and their clinical significance. Cell Rep. 2019;28(11):2866–2877.e5. - PubMed
-
- Reedijk AMJ, Klein K, Coebergh JWW, et al. Improved survival for children and young adolescents with acute myeloid leukemia: a Dutch study on incidence, survival and mortality. Leukemia. 2019;33(6):1349–1359. - PubMed
-
- Pollyea DA, Jordan CT. Therapeutic targeting of acute myeloid leukemia stem cells. Blood. 2017;129(12):1627–1635. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

