Novel determinants of NOTCH1 trafficking and signaling in breast epithelial cells
- PMID: 39663000
- PMCID: PMC11633778
- DOI: 10.26508/lsa.202403122
Novel determinants of NOTCH1 trafficking and signaling in breast epithelial cells
Abstract
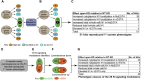
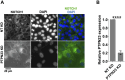
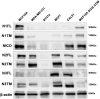
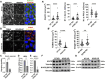
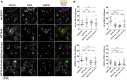
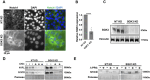
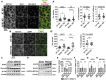
The evolutionarily conserved Notch signaling pathway controls cell-cell communication, enacting cell fate decisions during development and tissue homeostasis. Its dysregulation is associated with a wide range of diseases, including congenital disorders and cancers. Signaling outputs depend on maturation of Notch receptors and trafficking to the plasma membrane, endocytic uptake and sorting, lysosomal and proteasomal degradation, and ligand-dependent and independent proteolytic cleavages. We devised assays to follow quantitatively the trafficking and signaling of endogenous human NOTCH1 receptor in breast epithelial cells in culture. Based on such analyses, we executed a high-content screen of 2,749 human genes to identify new regulators of Notch that might be amenable to pharmacologic intervention. We uncovered 39 new NOTCH1 modulators for NOTCH1 trafficking and signaling. Among them, we find that PTPN23 and HCN2 act as positive NOTCH1 regulators by promoting endocytic trafficking and NOTCH1 maturation in the Golgi apparatus, respectively, whereas SGK3 serves as a negative regulator that can be modulated by pharmacologic inhibition. Our findings might be relevant in the search of new strategies to counteract pathologic Notch signaling.
© 2024 Kobia et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures