Examining the Influenza A Virus Sialic Acid Binding Preference Predictions of a Sequence-Based Convolutional Neural Network
- PMID: 39663148
- PMCID: PMC11634464
- DOI: 10.1111/irv.70044
Examining the Influenza A Virus Sialic Acid Binding Preference Predictions of a Sequence-Based Convolutional Neural Network
Abstract
Background: Though receptor binding specificity is well established as a contributor to host tropism and spillover potential of influenza A viruses, determining receptor binding preference of a specific virus still requires expensive and time-consuming laboratory analyses. In this study, we pilot a machine learning approach for prediction of binding preference.
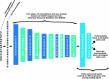
Methods: We trained a convolutional neural network to predict the α2,6-linked sialic acid preference of influenza A viruses given the hemagglutinin amino acid sequence. The model was evaluated with an independent test dataset to assess the standard performance metrics, the impact of missing data in the test sequences, and the prediction performance on novel subtypes. Further, features found to be important to the generation of predictions were tested via targeted mutagenesis of H9 and H16 proteins expressed on pseudoviruses.
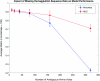
Results: The final model developed in this study produced predictions on a test dataset correctly 94% of the time and an area under the receiver operating characteristic curve of 0.93. The model tolerated about 10% missing test data without compromising accurate prediction performance. Predictions on novel subtypes revealed that the model can extrapolate feature relationships between subtypes when generating binding predictions. Finally, evaluation of the features important for model predictions helped identify positions that alter the sialic acid conformation preference of hemagglutinin proteins in practice.
Conclusions: Ultimately, our results provide support to this in silico approach to hemagglutinin receptor binding preference prediction. This work emphasizes the need for ongoing research efforts to produce tools that may aid future pandemic risk assessment.
Keywords: hemagglutinin; influenza; machine learning; receptor binding.
© 2024 The Author(s). Influenza and Other Respiratory Viruses published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Raman R., Venkataraman M., Ramakrishnan S., Lang W., Raguram S., and Sasisekharan R., “Advancing Glycomics: Implementation Strategies at the Consortium for Functional Glycomics,” Glycobiology 16, no. 5 (2006): 82R–90R. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

