Prediction of gene expression-based breast cancer proliferation scores from histopathology whole slide images using deep learning
- PMID: 39663527
- PMCID: PMC11633006
- DOI: 10.1186/s12885-024-13248-9
Prediction of gene expression-based breast cancer proliferation scores from histopathology whole slide images using deep learning
Abstract
Background: In breast cancer, several gene expression assays have been developed to provide a more personalised treatment. This study focuses on the prediction of two molecular proliferation signatures: an 11-gene proliferation score and the MKI67 proliferation marker gene. The aim was to assess whether these could be predicted from digital whole slide images (WSIs) using deep learning models.
Methods: WSIs and RNA-sequencing data from 819 invasive breast cancer patients were included for training, and models were evaluated on an internal test set of 172 cases as well as on 997 cases from a fully independent external test set. Two deep Convolutional Neural Network (CNN) models were optimised using WSIs and gene expression readouts from RNA-sequencing data of either the proliferation signature or the proliferation marker, and assessed using Spearman correlation (r). Prognostic performance was assessed through Cox proportional hazard modelling, estimating hazard ratios (HR).
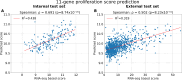
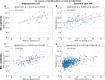
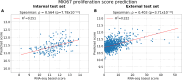
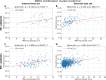
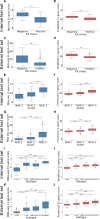
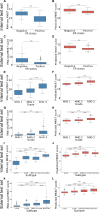
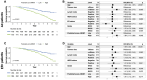
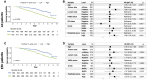
Results: Optimised CNNs successfully predicted the proliferation score and proliferation marker on the unseen internal test set (ρ = 0.691(p < 0.001) with R2 = 0.438, and ρ = 0.564 (p < 0.001) with R2 = 0.251 respectively) and on the external test set (ρ = 0.502 (p < 0.001) with R2 = 0.319, and ρ = 0.403 (p < 0.001) with R2 = 0.222 respectively). Patients with a high proliferation score or marker were significantly associated with a higher risk of recurrence or death in the external test set (HR = 1.65 (95% CI: 1.05-2.61) and HR = 1.84 (95% CI: 1.17-2.89), respectively).
Conclusions: The results from this study suggest that gene expression levels of proliferation scores can be predicted directly from breast cancer morphology in WSIs using CNNs and that the predictions provide prognostic information that could be used in research as well as in the clinical setting.
Keywords: Artificial intelligence; Breast cancer; Computational pathology; Gene expression; Proliferation.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The Clinseq study was approved by the Ethical Committee of Karolinska Institutet. Stockholm, Sweden, with reference 2013/1833-31/2. The SCAN-B study was approved by the Regional Ethical Review Board in Lund, Sweden (registration numbers 2009/658, 2010/383, 2012/58, 2013/459, 2014/521, 2015/277, 2016/541, 2016/742, 2016/944, 2018/267 and the Swedish Ethical Review Authority (registration numbers 2019-01252, 2024-02040-02). All patients provided written informed consent prior to enrollment. All analyses were performed in accordance with patient consent and ethical regulations and decisions. The study was further approved by the Swedish Ethical Review Authority (Etiksprovningsmyndigheten, Stockholm, Sweden, reference 2017/2106-31, with amendments 2018/1462-32 and 2019–02336). Consent for publication: Not applicable. Competing interests: MR is a cofounder and shareholder of Stratipath AB. YW is employed by Stratipath AB and holds employee stock options. AE, JVC and CB declare no financial or non-financial competing interests.
Figures
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians. 2021;71(3):209 – 49. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

