USP4 depletion-driven RAB7A ubiquitylation impairs autophagosome-lysosome fusion and aggravates periodontitis
- PMID: 39663592
- PMCID: PMC11925113
- DOI: 10.1080/15548627.2024.2429371
USP4 depletion-driven RAB7A ubiquitylation impairs autophagosome-lysosome fusion and aggravates periodontitis
Abstract
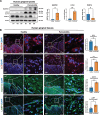
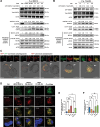
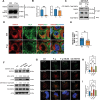
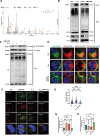
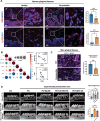
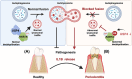
Periodontitis, a prevalent and chronic inflammatory disease, is intricately linked with macroautophagy/autophagy, which has a dual role in maintaining periodontal homeostasis. Despite its importance, the precise interplay between autophagy and periodontitis pathogenesis remains to be fully elucidated. In this study, our investigation revealed that the ubiquitination of RAB7A, mediated by reduced levels of the deubiquitinating enzyme USP4 (ubiquitin specific peptidase 4), disrupts normal lysosomal trafficking and autophagosome-lysosome fusion, thereby contributing significantly to periodontitis progression. Specifically, through genomic and histological analysis of clinical gingival samples, we observed a decreased RAB7A expression and impaired autophagic activity in periodontitis. This was further substantiated through experimental periodontitis mice, where RAB7A inactivation was shown to directly affect autophagy efficiency and drive periodontitis progression. Next, we explored the function of active RAB7A to promote lysosomal trafficking dynamics and autophagosome-lysosome fusion, which was inhibited by RAB7A ubiquitination in macrophages stimulated by Porphyromonas gingivalis (P. g.), one of the keystone pathogens of periodontitis. Last, by proteomics analysis, we revealed that the ubiquitination of RAB7A was mediated by USP4 and validated that upregulation of USP4 could attenuate periodontitis in vivo. In conclusion, these findings highlight the interaction between USP4 and RAB7A as a promising target for therapeutic intervention in managing periodontal diseases.Abbreviation: 3-MA: 3-methyladenine; Baf A1:bafilomycin A1; BECN1: beclin 1, autophagy related; CEJ-ABC: cementoenamel junctionto alveolar bone crest; IL1B/IL-1β: interleukin 1 beta; KD:knockdown; LPS: lipopolysaccharide; MOI: multiplicity of infection;OE: overexpression; P.g.: Porphyromonasgingivalis; RILP: Rabinteracting lysosomal protein; ScRNA-seq: single-cell RNA sequencing; SQSTM1/p62: sequestosome 1; S.s.: Streptococcus sanguinis; USP4:ubiquitin specific peptidase 4.
Keywords: Autophagy; GTP-RAB7A; IL1B; porphyromonas gingivalis; ubiquitination.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
